Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation - PubMed (original) (raw)
doi: 10.1128/JVI.00769-09.
Annamaria D'Aprile, Giovanni Quarato, Magdalena Sarasin-Filipowicz, Jérôme Gouttenoire, Rosella Scrima, Olga Cela, Domenico Boffoli, Markus H Heim, Darius Moradpour, Nazzareno Capitanio, Claudia Piccoli
Affiliations
- PMID: 19846525
- PMCID: PMC2798449
- DOI: 10.1128/JVI.00769-09
Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation
Maria Ripoli et al. J Virol. 2010 Jan.
Abstract
Hepatitis C virus (HCV) infection induces a state of oxidative stress by affecting mitochondrial-respiratory-chain activity. By using cell lines inducibly expressing different HCV constructs, we showed previously that viral-protein expression leads to severe impairment of mitochondrial oxidative phosphorylation and to major reliance on nonoxidative glucose metabolism. However, the bioenergetic competence of the induced cells was not compromised, indicating an efficient prosurvival adaptive response. Here, we show that HCV protein expression activates hypoxia-inducible factor 1 (HIF-1) by normoxic stabilization of its alpha subunit. In consequence, expression of HIF-controlled genes, including those coding for glycolytic enzymes, was significantly upregulated. Similar expression of HIF-controlled genes was observed in cell lines inducibly expressing subgenomic HCV constructs encoding either structural or nonstructural viral proteins. Stabilization and transcriptional activation of HIF-1alpha was confirmed in Huh-7.5 cells harboring cell culture-derived infectious HCV and in liver biopsy specimens from patients with chronic hepatitis C. The HCV-related HIF-1alpha stabilization was insensitive to antioxidant treatment. Mimicking an impairment of mitochondrial oxidative phosphorylation by treatment of inducible cell lines with oligomycin resulted in stabilization of HIF-1alpha. Similar results were obtained by treatment with pyruvate, indicating that accumulation of intermediate metabolites is sufficient to stabilize HIF-1alpha. These observations provide new insights into the pathogenesis of chronic hepatitis C and, possibly, the HCV-related development of hepatocellular carcinoma.
Figures
FIG. 1.
Effect of long-term HCV protein expression on the mitochondrial OXPHOS system in UHCVcon-57.3 cells. (A) Respiratory activity of intact UHCVcon-57.3 cells. Measurements of oxygen consumption were performed by high-resolution respirometry, as described in Materials and Methods. Repression of HCV protein expression is indicated as + Tet (i.e., medium supplemented with 1 μg/ml tetracycline); derepression of HCV proteins is indicated as − Tet (i.e., medium without tetracycline). The incubation time is also shown. End. resp., endogenous respiration; End. resp. + oligom., respiratory activity in the presence of oligomycin. The values represent averages of six independent cell preparations plus standard errors of the mean (SEM); the P values reported are versus the non-HCV-induced condition. (B) Mitochondrial OXPHOS complex activities. Specific activities of complex I (CI), CIII, CIV, and CV were measured by spectrophotometrical assays as specified in Materials and Methods. Empty bars, noninduced cells; light-gray and dark-gray bars, cells induced for 2 and 5 days, respectively. The activities of the complexes from induced cells were normalized to the values of control cells. The absolute activities were 3.60 nmol NADH/min/106 cells for CI; 3.43 nmol cytochrome c reduced/min/106 cells for CIII, 8.46 nmol cytochrome c oxidized/min/106 cells for CIV, and 6.09 nmol ATP/min/106 cells for CV. The values represent averages of four independent experiments plus SEM. The P value is also shown where significant. (C) Western blotting of HCV proteins in total cell lysates and mitochondrion-enriched fractions. Fifty micrograms of proteins was loaded on the gels in all cases. β-Actin, the β-subunit of the FoF1-ATP synthase (β-CV), and the Ca2+ pump ATPase (SERCA) were taken as cytosol, mitochondrial, and ER markers, respectively. The results are representative of two experiments. (D) Cell growth analysis. Light microscopy images of noninduced (+ Tet) cells and cells induced (− Tet) for 1, 2, and 5 days. Cell numbers are shown. The results are averages of five independent assays ± SEM; the statistical significance versus control cells is shown.
FIG. 2.
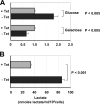
Effect of long-term HCV protein expression on cell bioenergetics. (A) Intracellular ATP content. The ATP contents of cell lysates were measured as described in Materials and Methods. Where indicated, glucose or galactose was the main carbon source in the DMEM base culture medium. Noninduced cells, + Tet (gray bars); 5-day-induced cells, − Tet (black bars). Each bar represents the average of five independent measurements plus the standard error of the mean (SEM) normalized to the ATP content measured in noninduced cells. The absolute mean values of ATP in noninduced cells were 5.9 and 8.4 nmol/mg protein in cells grown in glucose and galactose media, respectively. The statistical differences in ATP content between induced and noninduced cells are also shown. (B) Measurements of lactate in noninduced (gray bar) and 5-day-induced (black bar) UHCVcon-57.3 cells. The metabolite content was evaluated spectrophotometrically on cell culture media as described in Materials and Methods. The values reported in the histogram are the averages of three independent experiments plus SEM, along with the statistical difference.
FIG. 3.
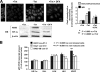
Effect of HCV protein expression on HIF-1α in U-2 OS-derived inducible cell lines. (A) HIF-1α protein expression. The three images at the top left are representative images (from four independent experiments) of immunofluorescence confocal microscopy analysis to detect HIF-1α. Noninduced UHCVcon-57.3 cells, +Tet; 5-day-induced cells, −Tet; noninduced cells treated with 10 mM desferroxiamine for 2 h, −Tet + DFX. The secondary fluorescein isothiocyanate-conjugated antibody (2nd-Ab-FITC), when added directly to the cell sample without the primary anti-HIF-1α, did not result in a detectable fluorescence signal (not shown). The inset in the image relative to induced cells shows a double immunodetection, in a parallel sample, of HIF-1α and the HCV NS5B protein. The latter was immunodetected using a secondary rhodamine-conjugated antibody (red fluorescence). Bars, 10 μm. A representative immunoblot (Western blot [WB]) analysis of HIF-1α and HCV NS5A protein expression on cell lysates under identical conditions is also shown. The histogram on the right displays the quantitative analysis of the immunofluorescence intensity (gray bars) assessed by averaging 20 to 30 single cells from at least 10 different optical fields; the mean values from four different cell preparations for each condition are shown, along with statistical analysis. The histogram also shows the result of densitometric analysis (black bars) of the WB HIF-1α band normalized to the internal β-actin control. The average value plus standard error of the mean (SEM) of three independent immunoblot assays is shown, along with statistical analysis. (B) Transcript levels of HIF-1-controlled genes. The transcript levels of HIF-1α, VHL, LDH-A, HK1, and VEGF genes were quantified by real-time RT-PCR as described in Materials and Methods. The analysis was carried out on U-2 OS-derived cell lines cultured for 5 days in the absence of tetracycline to express the entire HCV polyprotein (UHCVcon-57.3); the structural proteins, as well as p7 (UCp7con-9.10); or the nonstructural proteins 3 to 5B (UNS3-5Bcon-27). The expression level of each gene is shown as a percentage of that attained in noninduced cells and represents the average of three independent experiments plus SEM, along with statistical analysis.
FIG. 4.
Effects of GFP expression in U-2 OS-derived inducible cell lines. UGFP-9.22 cells were cultured in the presence (+) or absence (−) of tetracycline (Tet) for 5 days. (A) Confocal microscopy analysis showing GFP expression and mtΔΨ generation in mitochondria assessed by the probe TMRE. Bars, 10 μm. (B) Western blot (WB) of HIF-1α. The blot is representative of three independent experiments. (C) Transcript levels of HIF-1-controlled genes. The transcript levels of HIF-1α, VHL, LDH-A, HK1, and VEGF genes were quantified by real-time RT-PCR as described in Materials and Methods. The expression level of each gene is shown as a percentage of that attained in noninduced cells and represents the average of three independent experiments plus the standard error of the mean (P > 0.05 for all the genes).
FIG. 5.
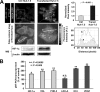
Effect of HCV protein expression on the activity of HIF-1 in transiently transfected Huh-7.5 cells. (A) HIF-1α immunofluorescence confocal microscopy analysis. Representative images of control and Jc1-GFP RNA-transfected Huh-7.5 cells are shown illustrating the merge from the rhodamine (R)-related HIF-1α immunodetection and the GFP-related fluorescence signal; the latter was used to track HCV-transfected cells (>95% of the cell population). Control Huh-7.5 cells were subjected to the transfection protocol but without HCV RNA. The time point after transfection was 72 h. see Materials and Methods for further details. Bars, 10 μm. Enlargements with false-color imaging of the original pictures are also shown to better visualize the nuclear localization of HIF-1α. On the right, a histogram of the HIF-1α-related fluorescence in GFP-positive transfected cells compared to control cells is shown. The fluorescence intensity was assessed by averaging 10 to 20 cells from each of at least 10 optical fields. The average of four independent experiments plus the standard error of the mean (SEM), along with statistical analysis, is shown. A density profile of the HIF-1α-related immunofluorescence signal is representatively shown along two lines crossing two selected control and transfected cells. A representative Western blot (WB) of HIF-1α on cell lysates of parallel samples is also shown. A.U., arbitrary units. (B) Effect of HCV protein expression on the transcript level of HIF-1-controlled genes in transfected Huh-7.5 cells. The transcript levels of HIF-1α, VHL, PHD2, LDH-A, HK1, and VEGF genes were quantified by real-time RT-PCR as described in Materials and Methods. The expression level of each gene is shown as a percentage of that attained in nontransfected control cells and represents the average of three independent experiments plus the standard error of the mean, along with statistical analysis.
FIG. 6.
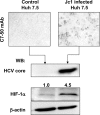
Stabilization of HIF-1α in HCV-infected Huh-7.5 cells. Huh-7.5 cells were infected with Jc1 HCV as described in Materials and Methods and analyzed 4 days after infection. (Top) Micrographs of cells stained for HCV core protein, as revealed by MAb C7-50 and a horseradish peroxidase-conjugated secondary antibody. (Bottom) Representative (out of three independent preparations) Western blots (WB) of cell lysates for HCV core, HIF-1α, and β-actin. Relative densitometric values for HIF-1α normalized to β-actin are indicated.
FIG. 7.
Effects of antioxidants and inhibitors on HCV-induced HIF-1 activation. (A) LSCM analysis of intracellular ROS production. Imaging of intracellular H2O2 was evaluated by DCF as specified in Materials and Methods. UHCVcon-57.3 cells were induced for 5 days. Where indicated, induced cells were treated for the entire induction time with either 20 mM NAC or 0.5 mM Tiron. Bars, 10 μm. The quantitative analysis of the DCF fluorescence from three different experiments is displayed as bars superimposed on the images; the fluorescence mean ± standard error of the mean (SEM), averaging the pixel intensities of 10 to 15 cell from at least 10 optical fields, is shown for each condition, along with statistical analysis. A representative Western blot (WB) of HIF-1α and HCV core on cell lysates of parallel samples is also shown. A.U., arbitrary units. (B) qRT-PCR assay of HIF-1α, VHL, LDH, HK1, and VEGF-A transcript levels. For experimental details, see Materials and Methods and the legend to Fig. 5. The light-gray and dark-gray bars refer to the effects of NAC and Tiron, respectively, under the experimental conditions of panel A. The level of each transcript was normalized to that of the noninduced cells, as in Fig. 5. The values shown are the ratios of the NAC- and Tiron-treated cells with respect to the induced untreated cells and represent the average of three independent experiments plus SEM. (C) Effects of ruthenium red (RR), dantrolene (Dantr.), wortmannin (Wortm.), and
l
-NNMA on the RT-PCR-assayed transcript levels of HIF-1α, VHL, LDH, HK1, and VEGF-A. UHCVcon-57.3 cells were induced for 5 days in the presence of each inhibitor at the following concentrations: 5 μM RR, 10 μM Dantr., 100 nM Wortm., and 1 mM
l
-NNMA. The transcript levels are expressed as in panel B and were averaged from two independent experiments.
FIG. 8.
Effects of antioxidants in transiently HCV-transfected Huh-7.5 cells. (A, left) ROS production in intact Huh-7.5 cells assessed by fluorimetry as described in Materials and Methods. The experimental traces refer to control and Jc1-GFP RNA-transfected Huh-7.5 cells (time point, 3 days after transfection) and were corrected point by point over the same time course for the recordings attained under identical conditions with cells treated with 20 mM NAC (comparable outcomes were obtained when corrections were made for cells treated with 0.5 mM Tiron). The rates of ROS generation in control and transfected Huh-7.5 cells without/with NAC or Tiron treatment are shown on the right. The values were calculated from the initial increase of the fluorescence following the addition of DCF-DA and are the average plus the standard error of the mean (SEM), along with statistical analysis, of recordings carried out in triplicate for each condition from three independent experiments. (B) Western blot of HIF-1α and HCV core on cell lysates of control and HCV-transfected Huh-7.5 cells, the latter without/with NAC or Tiron treatment. the results are representative of three independent experiments. (C) qRT-PCR assay of HIF-1α and VEGF-A transcript levels. For experimental details, see Materials and Methods and the legend to Fig. 5. The dark-gray and light-gray bars refer to the effects of NAC and Tiron, respectively, under the experimental conditions of panel A in transfected Huh-7.5 cells. The level of each transcript was normalized to that of the noninduced cells as in Fig. 5 and represents the average of three independent experiments plus SEM, along with statistical analysis.
FIG. 9.
Effects of oligomycin and pyruvate on HIF-1α stabilization in U-2 OS cells. (A, left) Immunofluorescence analysis of HIF-1α in noninduced UHCVcon-57.3 cells. For experimental details, see Materials and Methods and the legend to Fig. 3. Where indicated, cells were treated with 1 μM oligomycin or 2 mM pyruvate for 4 h. Bars, 10 μm. False-color images of the original pictures are shown to better visualize the nuclear localization of HIF-1α. The average fluorescence intensities from three independent experiments plus standard errors of the mean, along with statistical analysis, are represented in the histogram on the right. (B) Western blot (WB) of HIF-1α on lysates of cell samples treated as in panel A; the results are representative of three independent experiments. Oligo., oligomycin; Pyr., pyruvate.
FIG. 10.
Effects of oligomycin and pyruvate on HIF-1α stabilization in Huh-7.5 cells. (A, left) Immunofluorescence analysis of HIF-1α in Huh-7.5 cells. For experimental details, see Materials and Methods and the legend to Fig. 3. Where indicated, cells were treated with 1 μM oligomycin or 2 mM pyruvate for 4 h. Bars, 10 μm. The average fluorescence intensities from three independent experiments plus standard errors of the mean, along with statistical analysis, are represented in the histogram on the right. (B) Western blot (WB) of HIF-1α on lysates of cell samples treated as in panel A; the results are representative of three independent experiments. Oligo., oligomycin; Pyr., pyruvate.
FIG. 11.
Expression of HIF-1α-controlled genes in liver biopsy specimens from HCV-infected patients. Shown is qRT-PCR analysis of HIF-1α, VHL, LDH-A, HK1, and VEGF mRNAs in 14 healthy control liver biopsy samples (Controls) and 19 samples derived from patients with CHC (HCV). P values were determined by two-tailed Mann-Whitney tests.
FIG. 12.
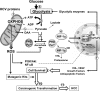
Proposed mechanism for the HCV-linked normoxic stabilization of HIF-1 and its pathogenic implications. The impairment of the mitochondrial OXPHOS system caused by HCV proteins (1) is suggested to induce a metabolic shift toward glycolysis. This persistent metabolic setting would cause accumulation of pyruvate and Krebs cycle intermediates (2). These are proposed to inhibit the PHDs (3), thereby stabilizing HIF-1α (4). Nuclear translocation of HIF-1α and transactivation of hypoxia-responding genes would upregulate the expression of glycolitic enzymes (5). Therefore, a positive feed-forward mechanism is activated (6). Moreover, as a “side effect,” other HIF-dependent angiogenetic and prosurvival factors are upregulated (7 and 8). These events, in combination with HCV protein expression-dependent ROS overproduction (9), may eventually lead to carcinogenic transformation (10 and 11). See the text for further details. OAA, oxalacetate.
Similar articles
- Impairment of HIF-1α-mediated metabolic adaption by NRF2-silencing in breast cancer cells.
Lee S, Hallis SP, Jung KA, Ryu D, Kwak MK. Lee S, et al. Redox Biol. 2019 Jun;24:101210. doi: 10.1016/j.redox.2019.101210. Epub 2019 May 2. Redox Biol. 2019. PMID: 31078780 Free PMC article. - Pyruvate dehydrogenase kinase regulates hepatitis C virus replication.
Jung GS, Jeon JH, Choi YK, Jang SY, Park SY, Kim SW, Byun JK, Kim MK, Lee S, Shin EC, Lee IK, Kang YN, Park KG. Jung GS, et al. Sci Rep. 2016 Jul 29;6:30846. doi: 10.1038/srep30846. Sci Rep. 2016. PMID: 27471054 Free PMC article. - Protective role of amantadine in mitochondrial dysfunction and oxidative stress mediated by hepatitis C virus protein expression.
Quarato G, Scrima R, Ripoli M, Agriesti F, Moradpour D, Capitanio N, Piccoli C. Quarato G, et al. Biochem Pharmacol. 2014 Jun 15;89(4):545-56. doi: 10.1016/j.bcp.2014.03.018. Epub 2014 Apr 12. Biochem Pharmacol. 2014. PMID: 24726442 - HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms.
Marín-Hernández A, Gallardo-Pérez JC, Ralph SJ, Rodríguez-Enríquez S, Moreno-Sánchez R. Marín-Hernández A, et al. Mini Rev Med Chem. 2009 Aug;9(9):1084-101. doi: 10.2174/138955709788922610. Mini Rev Med Chem. 2009. PMID: 19689405 Review. - Interplay between hypoxia inducible Factor-1 and mitochondria in cardiac diseases.
Mialet-Perez J, Belaidi E. Mialet-Perez J, et al. Free Radic Biol Med. 2024 Aug 20;221:13-22. doi: 10.1016/j.freeradbiomed.2024.04.239. Epub 2024 Apr 30. Free Radic Biol Med. 2024. PMID: 38697490 Review.
Cited by
- Hypoxic gene expression in chronic hepatitis B virus infected patients is not observed in state-of-the-art in vitro and mouse infection models.
Liu PJ, Harris JM, Marchi E, D'Arienzo V, Michler T, Wing PAC, Magri A, Ortega-Prieto AM, van de Klundert M, Wettengel J, Durantel D, Dorner M, Klenerman P, Protzer U, Giotis ES, McKeating JA. Liu PJ, et al. Sci Rep. 2020 Aug 24;10(1):14101. doi: 10.1038/s41598-020-70865-7. Sci Rep. 2020. PMID: 32839523 Free PMC article. - Quantitative proteomics of Sf21 cells during Baculovirus infection reveals progressive host proteome changes and its regulation by viral miRNA.
Nayyar N, Kaur I, Malhotra P, Bhatnagar RK. Nayyar N, et al. Sci Rep. 2017 Sep 7;7(1):10902. doi: 10.1038/s41598-017-10787-z. Sci Rep. 2017. PMID: 28883418 Free PMC article. - Differential regulation of cytotoxicity pathway discriminating between HIV, HCV mono- and co-infection identified by transcriptome profiling of PBMCs.
Wu JQ, Saksena MM, Soriano V, Vispo E, Saksena NK. Wu JQ, et al. Virol J. 2015 Jan 27;12:4. doi: 10.1186/s12985-014-0236-6. Virol J. 2015. PMID: 25623235 Free PMC article. - Untargeted Metabolomics Insights into Newborns with Congenital Zika Infection.
Nunes EDC, Filippis AMB, Pereira TDES, Faria NRDC, Salgado Á, Santos CS, Carvalho TCPX, Calcagno JI, Chalhoub FLL, Brown D, Giovanetti M, Alcantara LCJ, Barreto FK, de Siqueira IC, Canuto GAB. Nunes EDC, et al. Pathogens. 2021 Apr 13;10(4):468. doi: 10.3390/pathogens10040468. Pathogens. 2021. PMID: 33924291 Free PMC article. - Endothelial Cell Metabolism in Vascular Functions.
Filippini A, Tamagnone L, D'Alessio A. Filippini A, et al. Cancers (Basel). 2022 Apr 11;14(8):1929. doi: 10.3390/cancers14081929. Cancers (Basel). 2022. PMID: 35454836 Free PMC article. Review.
References
- Abdalla, M. Y., I. M. Ahmad, D. R. Spitz, W. N. Schmidt, and B. E. Britigan. 2005. Hepatitis C virus-core and nonstructural proteins lead to different effects on cellular antioxidant defenses. J. Med. Virol. 76:489-497. - PubMed
- Appel, N., T. Schaller, F. Penin, and R. Bartenschlager. 2006. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 281:9833-9836. - PubMed
- Brass, V., E. Bieck, R. Montserret, B. Wölk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. - PubMed
- Brune, B., and J. Zhou. 2007. Hypoxia-inducible factor-1 alpha under the control of nitric oxide. Methods Enzymol. 435:463-478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources