Dengue virus capsid protein usurps lipid droplets for viral particle formation - PubMed (original) (raw)
Dengue virus capsid protein usurps lipid droplets for viral particle formation
Marcelo M Samsa et al. PLoS Pathog. 2009 Oct.
Abstract
Dengue virus is responsible for the highest rates of disease and mortality among the members of the Flavivirus genus. Dengue epidemics are still occurring around the world, indicating an urgent need of prophylactic vaccines and antivirals. In recent years, a great deal has been learned about the mechanisms of dengue virus genome amplification. However, little is known about the process by which the capsid protein recruits the viral genome during encapsidation. Here, we found that the mature capsid protein in the cytoplasm of dengue virus infected cells accumulates on the surface of ER-derived organelles named lipid droplets. Mutagenesis analysis using infectious dengue virus clones has identified specific hydrophobic amino acids, located in the center of the capsid protein, as key elements for lipid droplet association. Substitutions of amino acid L50 or L54 in the capsid protein disrupted lipid droplet targeting and impaired viral particle formation. We also report that dengue virus infection increases the number of lipid droplets per cell, suggesting a link between lipid droplet metabolism and viral replication. In this regard, we found that pharmacological manipulation of the amount of lipid droplets in the cell can be a means to control dengue virus replication. In addition, we developed a novel genetic system to dissociate cis-acting RNA replication elements from the capsid coding sequence. Using this system, we found that mislocalization of a mutated capsid protein decreased viral RNA amplification. We propose that lipid droplets play multiple roles during the viral life cycle; they could sequester the viral capsid protein early during infection and provide a scaffold for genome encapsidation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
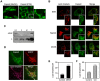
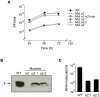
Figure 1. DENV infected cells accumulate the C protein around lipid droplets.
A. Nuclear and cytoplasmic distribution of C protein in DENV infected BHK cells. Cells were infected with DENV2 and analyzed by immunofluorescence using a polyclonal anti-C antibody. Cells were fixed with methanol (MeOH) or paraformaldehyde (PFA) as indicated on the top. B. The C protein is targeted to lipid droplets. BHK, HepG2, and C6/36 cells were infected with DENV2, fixed at 48 h post-infection, probed with anti-C antibodies and BODIPY for lipid droplets staining, and examined by confocal microscopy. C. Subcellular fractionation of LDs. DENV-infected cell lysates were fractionated into lipid droplets (LD), cytosol (C), and microsome (M) fractions by sucrose gradient centrifugation. A total cytoplasmic extract was also included (T). The samples were immunoblotted with anti-ADRP and anti-C antibodies. D. Co-localization of C and ADRP on LDs. DENV infected BHK cells were analyzed by immunofluorescence with anti-ADRP and anti-C antibodies, and stained with BODIPY. E. DENV infection increases the number of lipid droplets. The amount of lipid droplets in control or DENV infected BHK cells were determined. Cells were fixed 48 h post- infection, incubated in 1.5% of OsO4, and lipid bodies were enumerated by light microscopy in 50 consecutive cells in each slide in triplicates. The bars indicate the standard error of the mean (+/−SEM), (P<0.0002). F. Expression of C protein increases the number of lipid droplets. The amount of lipid droplets in control or C expressing BHK cells were determined as described above. The bars represent the standard error of the mean (P<0.0001).
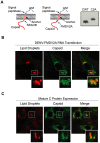
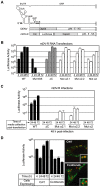
Figure 2. The C protein contains the structural determinants for LD targeting.
A. Schematic representation of the topology of the viral C and prM proteins on the ER membrane. The anchor peptide and the cleavage sites of the signal peptidase and viral NS3/2B proteases are indicated. The location of the FMDV2A protease replacing the NS3/2B site is shown in the scheme on the right. The western blot shows expression of the C protein in cytoplasmic extracts of cells transfected with a full length DENV RNA WT (Cwt) or the RNA including the FMDV2A site (C2A). B. The anchor peptide is dispensable for C accumulation on LDs. BHK cells transfected with the DENV-FMDV2A RNA were fixed and probed with antibodies against C and BODIPY to stain neutral lipids in LDs, as indicated on the top. C. Expression of the mature C protein in the absence of other viral components is sufficient for LD targeting. BHK cells were transfected with an expression plasmid that encode the mature form of DENV C protein. Twenty four h post-transfection cells were fixed and probed with anti-C antibodies followed by staining of lipid droplet.
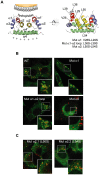
Figure 3. Amino acids within the α2 helix of C are necessary to direct the protein to LDs.
A. Ribbon diagram of the dimer structure of DENV C protein . The four α helices (α1 to α4) are indicated in each monomer. The hydrophobic cleft proposed to interact with membranes is also shown. On the right, the location of amino acids that were mutated in the DENV infectious clone is indicated in the structure (Mut α1, Mut α1–α2 loop, and Mut α2). B. Distribution of the C protein and lipid droplets in cells transfected with mutated DENV RNAs. BHK cells transfected with the WT or mutated RNAs containing the substitutions indicated in A were analyzed by immunofluorescence and confocal microscopy. The C protein and lipid droplets were localized by anti-C antibodies (green) and BODIPY (red), respectively. C. Amino acids L50 and L54 are necessary for targeting C to LDs. BHK cells transfected with DENV RNAs carrying the individual substitutions L50S (Mut α2.1) or L54S (Mut α2.2) were used to analyze the localization of the mutated C proteins and LDs as described above.
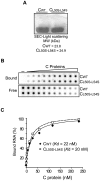
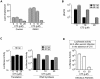
Figure 4. Biochemical properties of recombinant C protein with substitution L50S–L54S.
A. High expression levels and dimerization of CWT and CL50S–L54S. SDS-PAGE stained with coomassie blue showing similar expression levels of the recombinant proteins. The molecular mass obtained by size exclusion chromatography (SEC) and light scattering for both proteins are indicated. B. Interaction of CWT and CL50S–L54S with the DENV 5′UTR RNA probe monitored by filter binding assay. Uniformly 32P labeled RNA (0.1 nM) was incubated with increasing concentrations of the respective C protein. Bound indicates RNA-protein complexes retained in the nitrocellulose membrane and free denotes the unbound probes retained in the nylon membrane. The RNA probes bound and free in each membrane were visualized by PhosphoImaging. C. Quantification of the percentage of RNA probe bound was plotted as a function of C concentration and fitted using equation 1 (see Materials and methods). The dissociation constants K_ds_ are indicated inside the plot.
Figure 5. Targeting the C protein to LDs is necessary for DENV production.
A. The media of BHK cells transfected with DENV RNA WT or mutants (Mut α1, Mut α1–α2 loop, Mut α2, Mut α2.1, and Mut α2.2) were collected as a function of time post-transfection and used to quantify the amount of infectious particles by plaque assay in BHK cells. The plot indicates the plaque forming units per ml at different times post-transfection. B. The secreted enveloped protein E was analyzed in the supernatant of transfected cells by western blot as previously described . C. BHK cells were infected with a multiplicity of infection of 0.1 of WT, Mut α2.1, and Mut α2.2 viruses. The viral RNA was quantified by real time RT-PCR in the media obtained 24 h post-infection.
Figure 6. A new reporter virus that allows dissociation of cis-acting RNA elements from the capsid coding region confirms a role of L50 and L54 in DENV particle formation.
A. Construction of a novel monocistronic DENV reporter system. At the top, schematic representation of the cis-acting replication elements located at the 5′ end of the DENV genome. The promoter stem-loop A (SLA), the cyclization sequence upstream of the AUG (5′UAR), the replication element cHP, and the cyclization sequence 5′CS are indicated. In the middle, the corresponding region of DENV polyprotein is shown. At the bottom, a schematic representation of the monocistronic DENV reporter construct (mDV-R) showing the duplication of the cis-acting elements (CAE) and the location of the luciferase and the viral proteins. B. Translation and replication of mutant mDV-R RNAs. BHK cells were transfected with DENV RNAs corresponding to the mDV-R WT, Mut ΔC with the complete deletion of C coding sequence, Mut α2.1, Mut α2.2, Mut α2, and Mut NS5, which carries a mutation in the catalytic GDD motif of the viral polymerase. Luciferase activity was measured as a function of time for each RNA as indicated at the bottom. C. Mutations in the α2 helix of the C protein impair viral particle formation. The media of the transfected cells from the experiment shown in B was collected at the indicated times and used to infect fresh cells. Luciferase activity was measured 48 h post-infection for each virus as indicated at the bottom. D. A matured form of CL50SL54S protein expressed in BHK cells decreased the levels of DENV RNA synthesis. Immunofluorescence of BHK cells expressing the DENV CWT or CL50SL54S probed with anti C (green) and stained with Bodipy (red) for lipid droplets are shown in the right panel. The cells transfected with DV-R RNA WT were used to measure luciferase activity as a function of time, as indicated in the left panel.
Figure 7. Pharmacological inhibition of lipid droplets accumulation impairs DENV replication.
A. Effect of C75 on the amount of lipid droplets in BHK cells. The amount of lipid droplets was quantified in BHK cells treated with different concentrations of C75. Control or DENV infected BHK cells were used. B. Inhibition of DENV replication in cells treated with C75. The amount of infectious viral particles produced at 24 and 48 h post-infection in BHK cells were evaluated by plaque assays in control or C75 treated cells as indicated. Error bars indicate the SD of three independent experiments. C. Effect of C75 on each step of the replication of the mDV-R. Viral stocks of the reporter mDV-R were used to infect BHK cells in the presence and absence C75. Luciferase activity was evaluated at 10 h post-infection to evaluate entry and translation (left panel), and at 24 and 48 h to evaluate RNA synthesis (right panel). D. The production of infectious viral particles produced in the experiment described in C was evaluated by infecting fresh BHK cells in the absence of the inhibitor, and assessing the luciferase activity 48 h after infection.
Similar articles
- Uncoupling cis-Acting RNA elements from coding sequences revealed a requirement of the N-terminal region of dengue virus capsid protein in virus particle formation.
Samsa MM, Mondotte JA, Caramelo JJ, Gamarnik AV. Samsa MM, et al. J Virol. 2012 Jan;86(2):1046-58. doi: 10.1128/JVI.05431-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072762 Free PMC article. - Properties and Functions of the Dengue Virus Capsid Protein.
Byk LA, Gamarnik AV. Byk LA, et al. Annu Rev Virol. 2016 Sep 29;3(1):263-281. doi: 10.1146/annurev-virology-110615-042334. Epub 2016 Aug 3. Annu Rev Virol. 2016. PMID: 27501261 Free PMC article. Review. - Dengue and Zika virus capsid proteins bind to membranes and self-assemble into liquid droplets with nucleic acids.
Ambroggio EE, Costa Navarro GS, Pérez Socas LB, Bagatolli LA, Gamarnik AV. Ambroggio EE, et al. J Biol Chem. 2021 Sep;297(3):101059. doi: 10.1016/j.jbc.2021.101059. Epub 2021 Aug 8. J Biol Chem. 2021. PMID: 34375636 Free PMC article. - Dengue Virus Genome Uncoating Requires Ubiquitination.
Byk LA, Iglesias NG, De Maio FA, Gebhard LG, Rossi M, Gamarnik AV. Byk LA, et al. mBio. 2016 Jun 28;7(3):e00804-16. doi: 10.1128/mBio.00804-16. mBio. 2016. PMID: 27353759 Free PMC article. - Capsid protein is central to the birth of flavivirus particles.
Tan TY, Fibriansah G, Lok SM. Tan TY, et al. PLoS Pathog. 2020 May 28;16(5):e1008542. doi: 10.1371/journal.ppat.1008542. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32463839 Free PMC article. Review. No abstract available.
Cited by
- PTEN Lipid Phosphatase Activity Enhances Dengue Virus Production through Akt/FoxO1/Maf1 Signaling.
Liu B, Gao TT, Fu XY, Xu ZH, Ren H, Zhao P, Qi ZT, Qin ZL. Liu B, et al. Virol Sin. 2021 Jun;36(3):412-423. doi: 10.1007/s12250-020-00291-6. Epub 2020 Oct 12. Virol Sin. 2021. PMID: 33044659 Free PMC article. - Plasmid-based reverse genetics for probing phosphorylation-dependent viroplasm formation in rotaviruses.
Criglar JM, Crawford SE, Estes MK. Criglar JM, et al. Virus Res. 2021 Jan 2;291:198193. doi: 10.1016/j.virusres.2020.198193. Epub 2020 Oct 11. Virus Res. 2021. PMID: 33053412 Free PMC article. Review. - Dengue Virus Uses a Non-Canonical Function of the Host GBF1-Arf-COPI System for Capsid Protein Accumulation on Lipid Droplets.
Iglesias NG, Mondotte JA, Byk LA, De Maio FA, Samsa MM, Alvarez C, Gamarnik AV. Iglesias NG, et al. Traffic. 2015 Sep;16(9):962-77. doi: 10.1111/tra.12305. Epub 2015 Jun 29. Traffic. 2015. PMID: 26031340 Free PMC article. - Cyclin-Dependent Kinases 8 and 19 Regulate Host Cell Metabolism during Dengue Virus Serotype 2 Infection.
Butler M, Chotiwan N, Brewster CD, DiLisio JE, Ackart DF, Podell BK, Basaraba RJ, Perera R, Quackenbush SL, Rovnak J. Butler M, et al. Viruses. 2020 Jun 17;12(6):654. doi: 10.3390/v12060654. Viruses. 2020. PMID: 32560467 Free PMC article. - The internal architecture of leukocyte lipid body organelles captured by three-dimensional electron microscopy tomography.
Melo RC, Paganoti GF, Dvorak AM, Weller PF. Melo RC, et al. PLoS One. 2013;8(3):e59578. doi: 10.1371/journal.pone.0059578. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555714 Free PMC article.
References
- World Health Organization. Disease Outbreak News. 2009. http://www.who.int/csr/don/archive/disease/dengue_fever/en/
- Lindenbach BTH, Rice CM. Fields Virology. Philadelphia: Lippincott-Raven; 2007. Flaviviridae: the viruses and their replication. pp. 1101–1152.
- Nowak T, Farber PM, Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989;169:365–376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources