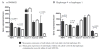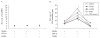Association of IRF5 polymorphisms with activation of the interferon alpha pathway - PubMed (original) (raw)
Association of IRF5 polymorphisms with activation of the interferon alpha pathway
Ornella J Rullo et al. Ann Rheum Dis. 2010 Mar.
Abstract
Objective: The genetic association of interferon regulatory factor 5 (IRF5) with systemic lupus erythematosus (SLE) susceptibility has been convincingly established. To gain understanding of the effect of IRF5 variation in individuals without SLE, a study was undertaken to examine whether such genetic variation predisposes to activation of the interferon alpha (IFNalpha) pathway.
Methods: Using a computer simulated approach, 14 single nucleotide polymorphisms (SNPs) and haplotypes of IRF5 were tested for association with mRNA expression levels of IRF5, IFNalpha and IFN-inducible genes and chemokines in lymphoblastoid cell lines (LCLs) from individuals of European (CEU), Han Chinese (CHB), Japanese (JPT) and Yoruba Nigerian (YRI) backgrounds. IFN-inducible gene expression was assessed in LCLs from children with SLE in the presence and absence of IFNalpha stimulation.
Results: The major alleles of IRF5 rs13242262 and rs2280714 were associated with increased IRF5 mRNA expression levels in the CEU, CHB+JPT and YRI samples. The minor allele of IRF5 rs10488631 was associated with increased IRF5, IFNalpha and IFN-inducible chemokine expression in CEU (p(c)=0.0005, 0.01 and 0.04, respectively). A haplotype containing these risk alleles of rs13242262, rs10488631 and rs2280714 was associated with increased IRF5, IFNalpha and IFN-inducible chemokine expression in CEU LCLs. In vitro studies showed specific activation of IFN-inducible genes in LCLs by IFNalpha.
Conclusions: SNPs of IRF5 in healthy individuals of a number of ethnic groups were associated with increased mRNA expression of IRF5. In European-derived individuals, an IRF5 haplotype was associated with increased IRF5, IFNalpha and IFN-inducible chemokine expression. Identifying individuals genetically predisposed to increased IFN-inducible gene and chemokine expression may allow early detection of risk for SLE.
Figures
Figure 1
Interferon regulatory factor 5 (IRF5) _g_ene structure and haplotypic architecture (A) Translated exons are shown as boxes. Single nucleotide polymorphisms (SNPs) used in the analysis have been encoded as: rs4728142 (SNP 1), rs752637 (SNP 2), rs3807306 (SNP 3), rs11761199 (SNP 4), rs6975315 (SNP 5), rs7808907 (SNP 6), rs1874328 (SNP 7), rs6970960 (SNP 8), rs6951628 (SNP 9), rs13242262 (SNP 10), rs1727172 (SNP 11), rs10488630 (SNP 12), rs10488631 (SNP 13) and rs2280714 (SNP 14). (B) Linkage disequilibrium (LD) r2 chart summarises the LD pattern in European (CEU), Han Chinese + Japanese (CHB+JPT) and Yoruba Nigerian (YRI) individuals as a plot. Darker grey represents regions of high pairwise r2 and white represents regions of low pairwise r2. The numbers in the boxes are the pairwise r2 values. TagSNPs are noted by asterisks. (C) The haplotypes across IRF5 constructed from 58 unrelated CEU individuals. The haplotypes are numbered on the left of each haplotype and haplotype frequencies are shown to the right of each haplotype. The SNP numbers across the top of the haplotypes correspond to those in the gene diagram above. The grey-boxed haplotype is the risk haplotype, defined by the risk alleles of SNPs 10, 13 and 14. The black-boxed haplotype is the non-risk haplotype.
Figure 2
Association of interferon regulatory factor 5 (IRF5) single nucleotide polymorphisms (SNPs) with increased IRF5 expression in three samples. IRF5 expression in European, Chinese/Japanese and Yoruba Nigerian individuals with or without risk alleles (homozygotes only) of (A) rs13242262 (SNP 10) and (B) rs2280714 (SNP 14). All p values withstand the Bonferroni correction method. SNP 14 is a proxy for the known functional SNP rs10954213, the polyadenylation variant site.
Figure 3
Mean mRNA expression (±SEM) of interferon regulatory factor 5 (IRF5) and interferon α (IFNα), and mean IFN or CMK scores (±SEM) of (A) individuals of European background (CEU) with or without the systemic lupus erythematosus (SLE)-associated risk allele of single nucleotide polymorphism (SNP) 13 (rs10488631) or (B) CEU individuals with the risk haplotype (H4, black) or the non-risk haplotype (H1, white). The risk haplotype is defined by the risk alleles of SNPs 10, 13 and 14 which are all located in the 3′ downstream region. All haplotypes, including haplotype 4 (H4, AAGGCTTACA) were previously deduced using Haplotyper 1.0 and are shown in figure 1C. p Values shown are those which remained significant after application of the Bonferroni correction.
Figure 4
Increased relative expression of interferon-inducible genes, but not housekeeping genes, was seen in lymphoblastoid cells (LCLs) after interferon α (IFNα) stimulation. Relative expression of genes in LCLs incubated with IFNα was measured based on values of RFLPO, a non-IFN-inducible housekeeping gene and compared with expression of cells incubated with IFNα plus recombinant B18R protein, an established inhibitor of IFN activation. (A) Relative expression of GAPDH, a non-IFN-inducible gene, after stimulation of cells with IFNα. (B) The relative expression of each IFN-inducible gene (ISG15, OAS1, OASL or MX-1), with or without incubation with IFNα, was combined from all seven cell lines. A summed total representing IFN score 1 is also presented in (B). Data points were connected by lines to illustrate the gene activation of a given gene. In both panels, data are expressed as mean ± SD (SD is not shown in (B) due to overlap between data sets). Each condition was done in duplicate and results shown were one of the two representative experiments. *p<0.05 (IFN score 1, p=0.0002; OASL, p=0.006; MX-1, p=0.03); †p<0.1 (ISG15, p=0.09; OAS1, p=0.06).
Similar articles
- Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients.
Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Niewold TB, et al. Arthritis Rheum. 2008 Aug;58(8):2481-7. doi: 10.1002/art.23613. Arthritis Rheum. 2008. PMID: 18668568 Free PMC article. - Association of IRF5 polymorphisms with increased risk for systemic lupus erythematosus in population of Crete, a southern-eastern European Greek island.
Zervou MI, Dorschner JM, Ghodke-Puranik Y, Boumpas DT, Niewold TB, Goulielmos GN. Zervou MI, et al. Gene. 2017 Apr 30;610:9-14. doi: 10.1016/j.gene.2017.02.003. Epub 2017 Feb 6. Gene. 2017. PMID: 28185859 - IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus.
Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, Thomas K, Walker D, Kamp S, Frost JM, Wong AK, Merrill JT, Alarcón-Riquelme ME, Tikly M, Ramsey-Goldman R, Reveille JD, Petri MA, Edberg JC, Kimberly RP, Alarcón GS, Kamen DL, Gilkeson GS, Vyse TJ, James JA, Gaffney PM, Moser KL, Crow MK, Harley JB. Niewold TB, et al. Ann Rheum Dis. 2012 Mar;71(3):463-8. doi: 10.1136/annrheumdis-2011-200463. Epub 2011 Nov 16. Ann Rheum Dis. 2012. PMID: 22088620 Free PMC article. - Interferon regulatory factors in human lupus pathogenesis.
Salloum R, Niewold TB. Salloum R, et al. Transl Res. 2011 Jun;157(6):326-31. doi: 10.1016/j.trsl.2011.01.006. Epub 2011 Feb 8. Transl Res. 2011. PMID: 21575916 Free PMC article. Review. - The genetics and biology of Irf5-mediated signaling in lupus.
Kozyrev SV, Alarcon-Riquelme ME. Kozyrev SV, et al. Autoimmunity. 2007 Dec;40(8):591-601. doi: 10.1080/08916930701510905. Autoimmunity. 2007. PMID: 18075793 Review.
Cited by
- Type I IFN response associated with mTOR activation in the TAFRO subtype of idiopathic multicentric Castleman disease.
Pai RL, Japp AS, Gonzalez M, Rasheed RF, Okumura M, Arenas D, Pierson SK, Powers V, Layman AAK, Kao C, Hakonarson H, van Rhee F, Betts MR, Kambayashi T, Fajgenbaum DC. Pai RL, et al. JCI Insight. 2020 May 7;5(9):e135031. doi: 10.1172/jci.insight.135031. JCI Insight. 2020. PMID: 32376796 Free PMC article. - Dysregulated Fcγ receptor IIa-induced cytokine production in dendritic cells of lupus nephritis patients.
Newling M, Fiechter RH, Sritharan L, Hoepel W, van Burgsteden JA, Hak AE, van Vollenhoven RF, van de Sande MGH, Baeten DLP, den Dunnen J. Newling M, et al. Clin Exp Immunol. 2020 Jan;199(1):39-49. doi: 10.1111/cei.13371. Epub 2019 Oct 7. Clin Exp Immunol. 2020. PMID: 31509231 Free PMC article. Clinical Trial. - IRF5 regulates unique subset of genes in dendritic cells during West Nile virus infection.
Chow KT, Driscoll C, Loo YM, Knoll M, Gale M Jr. Chow KT, et al. J Leukoc Biol. 2019 Feb;105(2):411-425. doi: 10.1002/JLB.MA0318-136RRR. Epub 2018 Nov 20. J Leukoc Biol. 2019. PMID: 30457675 Free PMC article. - Differential and Overlapping Immune Programs Regulated by IRF3 and IRF5 in Plasmacytoid Dendritic Cells.
Chow KT, Wilkins C, Narita M, Green R, Knoll M, Loo YM, Gale M Jr. Chow KT, et al. J Immunol. 2018 Nov 15;201(10):3036-3050. doi: 10.4049/jimmunol.1800221. Epub 2018 Oct 8. J Immunol. 2018. PMID: 30297339 Free PMC article. - Genetic variants of interferon regulatory factor 5 associated with chronic hepatitis B infection.
Sy BT, Hoan NX, Tong HV, Meyer CG, Toan NL, Song LH, Bock CT, Velavan TP. Sy BT, et al. World J Gastroenterol. 2018 Jan 14;24(2):248-256. doi: 10.3748/wjg.v24.i2.248. World J Gastroenterol. 2018. PMID: 29375210 Free PMC article.
References
- Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16:541–7. - PubMed
- Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol. 2006;18:676–82. - PubMed
- Pascual V, Banchereau J, Palucka AK. The central role of dendritic cells and interferon-alpha in SLE. Curr Opin Rheumatol. 2003;15:548–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR043814/AR/NIAMS NIH HHS/United States
- R01 AR043814-12/AR/NIAMS NIH HHS/United States
- R01 AR43814/AR/NIAMS NIH HHS/United States
- 5 K12 HC034510/HC/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical