Toward the use of genomics to study microevolutionary change in bacteria - PubMed (original) (raw)
Review
Toward the use of genomics to study microevolutionary change in bacteria
Daniel Falush. PLoS Genet. 2009 Oct.
Abstract
Bacteria evolve rapidly in response to the environment they encounter. Some environmental changes are experienced numerous times by bacteria from the same population, providing an opportunity to dissect the genetic basis of adaptive evolution. Here I discuss two examples in which the patterns of rapid change provide insight into medically important bacterial phenotypes, namely immune escape by Neisseria meningitidis and host specificity of Campylobacter jejuni. Genomic analysis of populations of bacteria from these species holds great promise but requires appropriate concepts and statistical tools.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures
Figure 1. Acquisition of new tbpB genes by subgroup III Neisseria meningitidis during epidemic spread.
Colours indicate the family of each tbpB allele, with red corresponding to family 4, green corresponding to family 1, and blue corresponding to family 3. The bars highlight the time frame, most common tbpB type, and geographical extent of each epidemic (in 1987, pilgrims from the Hajj pilgrimage briefly distributed the lineage worldwide). The circles correspond to variant genotypes. Small circles indicating that the variant allele was found in only one strain; large circles indicate it was found in between two and four strains.
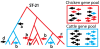
Figure 2. A schematic illustration of the evolution of the C. jejuni ST-21 clonal complex in cattle and chickens.
The common ancestor of the complex occurred in chickens (red). During evolution, the lineage occasionally switched to a cattle host (indicated by a blue branch) and sometimes back to chicken. The bacteria acquired DNA by homologous recombination from other C. jejuni in the same host. Since recombination is assumed to occur from donors within the same host, the gene pool is determined by the genomic composition of the strains that colonize each host. The gene pools are illustrated for two separate loci (right and left facing arrows) in chickens and cattle. The gene pools contain alleles whose frequencies occur at much higher frequency in one host than another (shown in colour) and others that did not (shown in black). The former are informative about the host in which the recombination event occurred, while the latter are not. The recombination event labelled a introduces the left facing black arrow gene from the cattle gene pool and is phylogenetically informative because it defines a lineage that is largely restricted to cattle. The five recombination events labelled b are not phylogenetically informative, since they only affect a single strain in the sample. These events are nevertheless informative because they introduce alleles that are characteristic of the host species. The event labelled c is both phylogenetically informative and characteristic of host. The event labelled d is noninformative.
Similar articles
- Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species.
Perrin A, Bonacorsi S, Carbonnelle E, Talibi D, Dessen P, Nassif X, Tinsley C. Perrin A, et al. Infect Immun. 2002 Dec;70(12):7063-72. doi: 10.1128/IAI.70.12.7063-7072.2002. Infect Immun. 2002. PMID: 12438387 Free PMC article. - Genomic Surveillance of 4CMenB Vaccine Antigenic Variants among Disease-Causing Neisseria meningitidis Isolates, United Kingdom, 2010-2016.
Rodrigues CMC, Lucidarme J, Borrow R, Smith A, Cameron JC, Moxon ER, Maiden MCJ. Rodrigues CMC, et al. Emerg Infect Dis. 2018 Apr;24(4):673-682. doi: 10.3201/eid2404.171480. Emerg Infect Dis. 2018. PMID: 29553330 Free PMC article. - Living in a changing environment: insights into host adaptation in Neisseria meningitidis from comparative genomics.
Schoen C, Joseph B, Claus H, Vogel U, Frosch M. Schoen C, et al. Int J Med Microbiol. 2007 Nov;297(7-8):601-13. doi: 10.1016/j.ijmm.2007.04.003. Epub 2007 Jun 14. Int J Med Microbiol. 2007. PMID: 17572149 Review. - Phylogenetic Methods for Genome-Wide Association Studies in Bacteria.
Didelot X. Didelot X. Methods Mol Biol. 2021;2242:205-220. doi: 10.1007/978-1-0716-1099-2_13. Methods Mol Biol. 2021. PMID: 33961226 - Neisseria genomics: current status and future perspectives.
Harrison OB, Schoen C, Retchless AC, Wang X, Jolley KA, Bray JE, Maiden MCJ. Harrison OB, et al. Pathog Dis. 2017 Aug 31;75(6):ftx060. doi: 10.1093/femspd/ftx060. Pathog Dis. 2017. PMID: 28591853 Free PMC article. Review.
Cited by
- Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa.
Tibayrenc M, Ayala FJ. Tibayrenc M, et al. Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):E3305-13. doi: 10.1073/pnas.1212452109. Epub 2012 Sep 4. Proc Natl Acad Sci U S A. 2012. PMID: 22949662 Free PMC article. - Molecular genomic approaches to infectious diseases in resource-limited settings.
Coloma J, Harris E. Coloma J, et al. PLoS Med. 2009 Oct;6(10):e1000142. doi: 10.1371/journal.pmed.1000142. Epub 2009 Oct 26. PLoS Med. 2009. PMID: 19855820 Free PMC article. - The Metallochaperone Encoding Gene hypA Is Widely Distributed among Pathogenic Aeromonas spp. and Its Expression Is Increased under Acidic pH and within Macrophages.
Fernández-Bravo A, López-Fernández L, Figueras MJ. Fernández-Bravo A, et al. Microorganisms. 2019 Oct 2;7(10):415. doi: 10.3390/microorganisms7100415. Microorganisms. 2019. PMID: 31581740 Free PMC article. - Impact of recombination on bacterial evolution.
Didelot X, Maiden MC. Didelot X, et al. Trends Microbiol. 2010 Jul;18(7):315-22. doi: 10.1016/j.tim.2010.04.002. Epub 2010 May 6. Trends Microbiol. 2010. PMID: 20452218 Free PMC article. Review. - Application of high-throughput genome sequencing to intrapathovar variation in Pseudomonas syringae.
Studholme DJ. Studholme DJ. Mol Plant Pathol. 2011 Oct;12(8):829-38. doi: 10.1111/j.1364-3703.2011.00713.x. Epub 2011 Apr 1. Mol Plant Pathol. 2011. PMID: 21726380 Free PMC article. Review.
References
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. - PubMed
- Weiss KM, Terwilliger JD. How many diseases does it take to map a gene with SNPs? Nat Genet. 2000;26:151–157. - PubMed
- Fabre M, Koeck JL, Le Fleche P, Simon F, Herve V, et al. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canettii” strains indicates that the M. tuberculosis complex is a recently emerged clone of “M. canettii”. J Clin Microbiol. 2004;42:3248–3255. - PMC - PubMed
- Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources