PDBe: Protein Data Bank in Europe - PubMed (original) (raw)
. 2010 Jan;38(Database issue):D308-17.
doi: 10.1093/nar/gkp916. Epub 2009 Oct 25.
C Best, B Beuth, C H Boutselakis, N Cobley, A W Sousa Da Silva, D Dimitropoulos, A Golovin, M Hirshberg, M John, E B Krissinel, R Newman, T Oldfield, A Pajon, C J Penkett, J Pineda-Castillo, G Sahni, S Sen, R Slowley, A Suarez-Uruena, J Swaminathan, G van Ginkel, W F Vranken, K Henrick, G J Kleywegt
Affiliations
- PMID: 19858099
- PMCID: PMC2808887
- DOI: 10.1093/nar/gkp916
PDBe: Protein Data Bank in Europe
S Velankar et al. Nucleic Acids Res. 2010 Jan.
Abstract
The Protein Data Bank in Europe (PDBe) (http://www.ebi.ac.uk/pdbe/) is actively working with its Worldwide Protein Data Bank partners to enhance the quality and consistency of the international archive of bio-macromolecular structure data, the Protein Data Bank (PDB). PDBe also works closely with its collaborators at the European Bioinformatics Institute and the scientific community around the world to enhance its databases and services by adding curated and actively maintained derived data to the existing structural data in the PDB. We have developed a new database infrastructure based on the remediated PDB archive data and a specially designed database for storing information on interactions between proteins and bound molecules. The group has developed new services that allow users to carry out simple textual queries or more complex 3D structure-based queries. The newly designed 'PDBeView Atlas pages' provide an overview of an individual PDB entry in a user-friendly layout and serve as a starting point to further explore the information available in the PDBe database. PDBe's active involvement with the X-ray crystallography, Nuclear Magnetic Resonance spectroscopy and cryo-Electron Microscopy communities have resulted in improved tools for structure deposition and analysis.
Figures
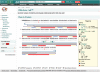
Figure 1.
Example of an Atlas page, in this case for PDB entry 1E9F. The menus on the left-hand side enable navigation between different areas of information as well as links to other resources and downloadable files. The main panel on the right displays the sequence annotated with secondary structure information from various other databases (Uniprot, CATH, Pfam and SCOP).
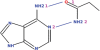
Figure 2.
Example of a graphically defined query that can be submitted to PDBeMotif.
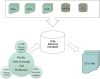
Figure 3.
Schematic overview of the process by which SIFTS files are generated (see text for details).
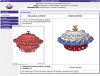
Figure 4.
The new EMViewer 3D visualization Java applet is available on the EMDB Atlas pages and allows interactive generation of isosurface representations. The map shown here is of the Borrelia flagellar motor, accession number EMDB-1644 (Authors: Kudryashev, M., Cyrklaff, M., Wallich, R., Baumeister, W., Frischknecht, F.).
Similar articles
- PDBe: Protein Data Bank in Europe.
Velankar S, Alhroub Y, Alili A, Best C, Boutselakis HC, Caboche S, Conroy MJ, Dana JM, van Ginkel G, Golovin A, Gore SP, Gutmanas A, Haslam P, Hirshberg M, John M, Lagerstedt I, Mir S, Newman LE, Oldfield TJ, Penkett CJ, Pineda-Castillo J, Rinaldi L, Sahni G, Sawka G, Sen S, Slowley R, Sousa da Silva AW, Suarez-Uruena A, Swaminathan GJ, Symmons MF, Vranken WF, Wainwright M, Kleywegt GJ. Velankar S, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D402-10. doi: 10.1093/nar/gkq985. Epub 2010 Nov 2. Nucleic Acids Res. 2011. PMID: 21045060 Free PMC article. - PDBe: Protein Data Bank in Europe.
Gutmanas A, Alhroub Y, Battle GM, Berrisford JM, Bochet E, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Gore SP, Haslam P, Hatherley R, Hendrickx PM, Hirshberg M, Lagerstedt I, Mir S, Mukhopadhyay A, Oldfield TJ, Patwardhan A, Rinaldi L, Sahni G, Sanz-García E, Sen S, Slowley RA, Velankar S, Wainwright ME, Kleywegt GJ. Gutmanas A, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D285-91. doi: 10.1093/nar/gkt1180. Epub 2013 Nov 27. Nucleic Acids Res. 2014. PMID: 24288376 Free PMC article. - PDBe: towards reusable data delivery infrastructure at protein data bank in Europe.
Mir S, Alhroub Y, Anyango S, Armstrong DR, Berrisford JM, Clark AR, Conroy MJ, Dana JM, Deshpande M, Gupta D, Gutmanas A, Haslam P, Mak L, Mukhopadhyay A, Nadzirin N, Paysan-Lafosse T, Sehnal D, Sen S, Smart OS, Varadi M, Kleywegt GJ, Velankar S. Mir S, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D486-D492. doi: 10.1093/nar/gkx1070. Nucleic Acids Res. 2018. PMID: 29126160 Free PMC article. - Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive.
Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S. Burley SK, et al. Methods Mol Biol. 2017;1607:627-641. doi: 10.1007/978-1-4939-7000-1_26. Methods Mol Biol. 2017. PMID: 28573592 Free PMC article. Review. - The Protein Data Bank Archive.
Velankar S, Burley SK, Kurisu G, Hoch JC, Markley JL. Velankar S, et al. Methods Mol Biol. 2021;2305:3-21. doi: 10.1007/978-1-0716-1406-8_1. Methods Mol Biol. 2021. PMID: 33950382 Review.
Cited by
- PCDDB: the Protein Circular Dichroism Data Bank, a repository for circular dichroism spectral and metadata.
Whitmore L, Woollett B, Miles AJ, Klose DP, Janes RW, Wallace BA. Whitmore L, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D480-6. doi: 10.1093/nar/gkq1026. Epub 2010 Nov 11. Nucleic Acids Res. 2011. PMID: 21071417 Free PMC article. - Modeling Structural Constraints on Protein Evolution via Side-Chain Conformational States.
Perron U, Kozlov AM, Stamatakis A, Goldman N, Moal IH. Perron U, et al. Mol Biol Evol. 2019 Sep 1;36(9):2086-2103. doi: 10.1093/molbev/msz122. Mol Biol Evol. 2019. PMID: 31114882 Free PMC article. - The Protein Data Bank in Europe (PDBe): bringing structure to biology.
Velankar S, Kleywegt GJ. Velankar S, et al. Acta Crystallogr D Biol Crystallogr. 2011 Apr;67(Pt 4):324-30. doi: 10.1107/S090744491004117X. Epub 2011 Mar 18. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21460450 Free PMC article. - Biochemical and structural characterization reveals Rv3400 codes for β-phosphoglucomutase in Mycobacterium tuberculosis.
Singh L, Karthikeyan S, Thakur KG. Singh L, et al. Protein Sci. 2024 Apr;33(4):e4943. doi: 10.1002/pro.4943. Protein Sci. 2024. PMID: 38501428 - Detection and analysis of unusual features in the structural model and structure-factor data of a birch pollen allergen.
Rupp B. Rupp B. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Apr 1;68(Pt 4):366-76. doi: 10.1107/S1744309112008421. Epub 2012 Mar 31. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22505400 Free PMC article.
References
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/E007511/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM079429/GM/NIGMS NIH HHS/United States
- BB/G022577/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- GR062025MA/WT_/Wellcome Trust/United Kingdom
- 088944/WT_/Wellcome Trust/United Kingdom