A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays - PubMed (original) (raw)
A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays
Danette D Hartzell et al. BMC Genomics. 2009.
Abstract
Background: Regulation of gene expression is essential for normal development and cellular growth. Transcriptional events are tightly controlled both spatially and temporally by specific DNA-protein interactions. In this study we finely map the genome-wide targets of the CREB protein across all known and predicted human promoters, and characterize the functional consequences of a subset of these binding events using high-throughput reporter assays. To measure CREB binding, we used HaloCHIP, an antibody-free alternative to the ChIP method that utilizes the HaloTag fusion protein, and also high-throughput promoter-luciferase reporter assays, which provide rapid and quantitative screening of promoters for transcriptional activation or repression in living cells.
Results: In analysis of CREB genome-wide binding events using a comprehensive DNA microarray of human promoters, we observe for the first time that CREB has a strong preference for binding at bidirectional promoters and unlike unidirectional promoters, these binding events often occur downstream of transcription start sites. Comparison between HaloCHIP-chip and ChIP-chip data reveal this to be true for both methodologies, indicating it is not a bias of the technology chosen. Transcriptional data obtained from promoter-luciferase reporter arrays also show an unprecedented, high level of activation of CREB-bound promoters in the presence of the co-activator protein TORC1.
Conclusion: These data suggest for the first time that TORC1 provides directional information when CREB is bound at bidirectional promoters and possible pausing of the CREB protein after initial transcriptional activation. Also, this combined approach demonstrates the ability to more broadly characterize CREB protein-DNA interactions wherein not only DNA binding sites are discovered, but also the potential of the promoter sequence to respond to CREB is evaluated.
Figures
Figure 1
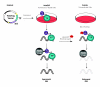
Schematic of the HaloCHIP process. The HaloCHIP process is initiated by cloning a desired DNA binding protein-of-interest, i.e. a transcription factor (TF) into a HaloTag (HT) fusion mammalian expression vector. For the experimental HaloCHIP sample, the HaloTag fusion protein is expressed in the desired cell line and then crosslinked to DNA in vivo with formaldehyde. Treated cells are lysed, sonicated to shear the chromatin, and incubated with HaloLink resin, which directly and covalently captures all crosslinked HaloTag-fusion complexes. The resin is stringently washed to remove non-specific proteins and DNA, and captured regions of DNA are released by reversal of the crosslinks. The resultant DNA can be further purified for downstream analysis. Two possible controls, which provide an estimate of background DNA capture, are recommended for the HaloCHIP process. The first, referred to in the text as the untransfected control, involves the use of untransfected cells which are processed in parallel with the HaloCHIP experimental sample. The second, referred to as the blocking ligand control, is generated by splitting the cell lysate equally into separate tubes prior to incubation with the HaloLink resin and incubating the control sample only with a fluorescent HaloCHIP blocking ligand, preventing interaction of the HaloTag complexes with the HaloLink resin during the subsequent incubation step.
Figure 2
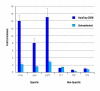
Specific DNA binding of HaloTag-CREB in vivo. HaloCHIP experiments were performed in triplicates, on HeLa cells transiently expressing HaloTag-CREB or untransfected as a control. Resulting DNA from both the HaloTag-CREB and Untransfected control sample was amplified and analyzed using Plexor quantitative PCR. Total amounts of DNA for both samples were calculated for three promoters which CREB is known to bind [28], Fos, Jun, and p27, as well as three control sequences, C1, C2, and C3, which do not contain CRE consensus binding sites. Depicted in dark blue is the fold enrichment of each CREB-specific promoter over the average amount of the three control promoters for the HaloTag-CREB HaloCHIP experimental sample. In light blue is the identical calculation for the Untransfected HaloCHIP control sample.
Figure 3
Schematic of the HaloCHIP-chip microarray experiment design. HaloCHIP DNA (10-50 ng) obtained for both the experimental HaloTag-CREB and untransfected control sample was purified and amplified to a concentration of 1-10 μg using the whole genome amplification (WGA) method (Sigma) [46]. The HaloTag-CREB amplified sample was labelled with Cy5 (green) and the untransfected control sample with Cy3 (red), then hybridized to a custom DNA oligo microarray manufactured by Roche NimbleGen. The oligo array was designed to cover on average a 1.8 kb region of 27,661 human promoter regions that contain 33,255 TSS predicted by SwitchGear Genomics. To obtain coverage of each promoter, an average of fourteen 50mer single stranded DNA probes, shown in purple, per promoter were used, with an average spacing of 131 bp per probe.
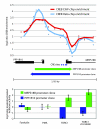
Figure 4
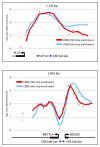
High-resolution analysis of CREB binding at uni-directional and bi-directional promoters. Depicted are CREB binding data from two representative promoter regions, the length (bp) of each is indicated above the graph. The log2 ratio for each probe spanning the promoter region was determined for the CREB ChIP-chip (plotted in red) or HaloCHIP-chip (plotted in blue) data. Positions of transcription start sites (TSS) are shown with arrows indicating the direction of transcription with length of exons in blocks and introns drawn as lines. Positions of full and half CRE sites relative to the location within the promoter region are indicated below by blue dots. A. Example of CREB binding to a unidirectional promoter, ALS2, where peak binding is observed upstream of the TSS and localized to the CRE consensus sites. B. Example of CREB binding to a bidirectional promoter, METTL4 and NDC80, where peak enrichment is located downstream of the TSSs, but the peak enrichment does not localize to CRE consensus sites.
Figure 5
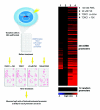
High-throughput reporter assays of CREB-bound promoters. A. Experimental design of high-throughput reporter assays. A schematic showing the high-throughput reporter assay experimental design. Promoters are fused to luciferase, transfected in a desired cell line, and stimulated under different conditions. Luciferase activity is measure from uninduced and induced samples and the log2 ratio of these differences is calculated. B. Heatmap of inducible promoter activity. A total of 235 promoters, chosen from CREB ChIP-chip and HaloCHIP-chip data, along with twelve negative control promoters were fused to the luciferase gene, transfected into HeLa cells, and treated with stimulants; forskolin (FSK), PMA, or co-transfected with a transcriptional co-activator, TORC1 +/- FSK. Each box represents the log2 ratio of induced/untreated for each promoter in each condition where the intensity of red is proportional to the strongest induction and the intensity of blue is proportional to the strongest repression. The presence of TORC1 by co-transfection shows the highest number of promoters induced to the highest degree. The box of 12 control promoters at the bottom of the panel are random promoters from the genome and show very little inducible activity in any of the conditions tested.
Figure 6
CREB binding and promoter activity at a bidirectional promoter showing opposite induction patterns in the presence of TORC1. A. CREB binding to PPP1R10/MRPS18B bidirectional promoter. Identical to Figure 4, the log2 ratio for each probe spanning this bidirectional promoter was determined for the CREB ChIP-chip (plotted in red) or HaloCHIP-chip (plotted in blue) data. Spacing between probes is approximately 131 bp. Positions of transcription start sites (TSS) are shown with arrows indicating the direction of transcription with lengths of exons in blocks and introns drawn as lines. Positions of full CRE sites are indicated below by blue dots. Also shown is the region of each cloned promoter fragments, indicated by the green and blue arrows, used in the luciferase assay shown below in panel B. The arrows indicate the direction in which the cloned fragments were tested in the luciferase assay. B. The log2 ratio of treated/untreated luciferase activity is plotted for each promoter fragment in each condition indicated. The MRPS18B promoter (in green) shows significant inducible activity in the presence of TORC1 with and without FSK, while the PPP1R10 promoter (in blue) shows significant repression in the presence of TORC1 with and without FSK.
Similar articles
- cAMP response element binding protein (CREB) activates transcription via two distinct genetic elements of the human glucose-6-phosphatase gene.
Thiel G, Al Sarraj J, Stefano L. Thiel G, et al. BMC Mol Biol. 2005 Jan 19;6:2. doi: 10.1186/1471-2199-6-2. BMC Mol Biol. 2005. PMID: 15659240 Free PMC article. - Anaerobic fluorescent reporters for cell identification, microbial cell biology and high-throughput screening of microbiota and genomic libraries.
Streett H, Charubin K, Papoutsakis ET. Streett H, et al. Curr Opin Biotechnol. 2021 Oct;71:151-163. doi: 10.1016/j.copbio.2021.07.005. Epub 2021 Aug 7. Curr Opin Biotechnol. 2021. PMID: 34375813 Review. - Life on a microarray: assessing live cell functions in a microarray format.
Papp K, Szittner Z, Prechl J. Papp K, et al. Cell Mol Life Sci. 2012 Aug;69(16):2717-25. doi: 10.1007/s00018-012-0947-z. Epub 2012 Mar 4. Cell Mol Life Sci. 2012. PMID: 22391673 Free PMC article. Review.
Cited by
- The HaloTag: Improving Soluble Expression and Applications in Protein Functional Analysis.
N Peterson S, Kwon K. N Peterson S, et al. Curr Chem Genomics. 2012;6:8-17. doi: 10.2174/1875397301206010008. Epub 2012 Sep 20. Curr Chem Genomics. 2012. PMID: 23115610 Free PMC article. - Linking CREB function with altered metabolism in murine fibroblast-based model cell lines.
Steven A, Leisz S, Wickenhauser C, Schulz K, Mougiakakos D, Kiessling R, Denkert C, Seliger B. Steven A, et al. Oncotarget. 2017 Oct 27;8(57):97439-97463. doi: 10.18632/oncotarget.22135. eCollection 2017 Nov 14. Oncotarget. 2017. PMID: 29228623 Free PMC article. - HaloTag, a Platform Technology for Protein Analysis.
Urh M, Rosenberg M. Urh M, et al. Curr Chem Genomics. 2012;6:72-8. doi: 10.2174/1875397301206010072. Epub 2012 Dec 5. Curr Chem Genomics. 2012. PMID: 23213345 Free PMC article. - Zinc-mediated activation of CREB pathway in proliferation of pulmonary artery smooth muscle cells in pulmonary hypertension.
Xiao G, Lian G, Wang T, Chen W, Zhuang W, Luo L, Wang H, Xie L. Xiao G, et al. Cell Commun Signal. 2021 Oct 11;19(1):103. doi: 10.1186/s12964-021-00779-y. Cell Commun Signal. 2021. PMID: 34635097 Free PMC article. - A novel regulatory circuit in base excision repair involving AP endonuclease 1, Creb1 and DNA polymerase beta.
Pei DS, Yang XJ, Liu W, Guikema JE, Schrader CE, Strauss PR. Pei DS, et al. Nucleic Acids Res. 2011 Apr;39(8):3156-65. doi: 10.1093/nar/gkq1142. Epub 2010 Dec 20. Nucleic Acids Res. 2011. PMID: 21172930 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases