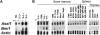Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia - PubMed (original) (raw)
Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia
Cynthia L Fisher et al. Blood. 2010.
Abstract
The Additional sex combs like 1 (Asxl1) gene is 1 of 3 mammalian homologs of the Additional sex combs (Asx) gene of Drosophila. Asx is unusual because it is required to maintain both activation and silencing of Hox genes in flies and mice. Asxl proteins are characterized by an amino terminal homology domain, by interaction domains for nuclear receptors, and by a C-terminal plant homeodomain protein-protein interaction domain. A recent study of patients with myelodysplastic syndrome (MDS) and chronic myelomonocytic leukemia (CMML) revealed a high incidence of truncation mutations that would delete the PHD domain of ASXL1. Here, we show that Asxl1 is expressed in all hematopoietic cell fractions analyzed. Asxl1 knockout mice exhibit defects in frequency of differentiation of lymphoid and myeloid progenitors, but not in multipotent progenitors. We do not detect effects on hematopoietic stem cells, or in peripheral blood. Notably, we do not detect severe myelodysplastic phenotypes or leukemia in this loss-of-function model. We conclude that Asxl1 is needed for normal hematopoiesis. The mild phenotypes observed may be because other Asxl genes have redundant function with Asxl1, or alternatively, MDS or oncogenic phenotypes may result from gain-of-function Asxl mutations caused by genomic amplification, gene fusion, or truncation of Asxl1.
Figures
Figure 1
Generation of Asxl1 mutant mice. (A) Schematic representation of the conserved ASXH and PHD domains and NR sequence motifs in ASXL1 homologs is shown. Numbers above the diagram show the first and last amino acids that define the ASXH and PHD domains. Mutations causing ASXL1 truncations found in MDS and CMML patients lie between amino acids 596 and 1457, all within exon 12. The arrowhead shows the site of insertion of the neomycin (neo) transgene at amino acid 90. (B) Diagram of part of the Asxl1 locus showing exons 5 to 13 indicated as black boxes. The PGKneo expression cassette (large white box) was inserted into the _Xba_I site in exon 5 by a replacement gene-targeting approach and was used for positive selection of clones. Position of relevant restriction sites (C indicates _Cla_I; E, _Eco_RI; H, _Hin_dIII; and X, _Xba_I), the location of external probe (bar below the E-H fragment shown at the left), and PCR primers (small arrows) are indicated. The location of the 5′ (_Hin_dIII site) and 3′ (_Cla_I site) ends of the flanking genomic homology arms in the targeting vector is denoted in larger bold font. (C) Southern blot analysis of genomic DNA isolated from newborn offspring of an Asxl1tm1Bc intercross after digestion with _Eco_RI and hybridized with an external probe shown in panel B. This probe detects a 7.3-kb fragment from the wild-type allele (+/+), and a 3.9-kb fragment from the targeted allele (m/m). (D) Multiple primer PCR analysis of liver genomic DNA of E18.5 embryos from an Asxl1tm1Bc intercross. Primers P2 and P5 amplify a 253-bp fragment of the wild-type allele, whereas primers P3 and P5 amplify a 426-bp fragment of the targeted allele. (E) Northern blot analysis of poly(A)+RNA from pooled tissue of neonate wild-type and Asxl1tm1Bc mice using probes for Asxl1 (left panel) and neomycin (right panel). (F) RT-PCR analysis of Asxl1tm1Bc mutant neonate tissue. RT-PCR of total RNA from pooled tissue of individual newborn Asxl1+/+ (+/+) and Asxl1tm1Bc/tm1Bc (m/m) mice. Primer from exon 1 and primer P5 (in exon 5) amplify an approximately 400-bp product in Asxl1+/+ samples, and a 2.2-kb product in Asxl1tm1Bc/tm1Bc (left panel). Primer from exon 1 and primer P4 (within neo) amplify a 300-bp product from the Asxl1tm1Bc/tm1Bc samples, whereas no amplification occurs in the Asxl1+/+ samples, as expected (right panel).
Figure 2
Asxl1 is ubiquitously expressed in hematopoietic cells. Southern blot analysis of Asxl1, Bmi1, and actin gene expression in total amplified cDNAs from FACS-sorted hematopoietic cell populations from fetal liver and adult bone marrow, spleen, and thymus. Total cDNAs were amplified using RT-PCR from 104 sorted cells, and each sample with or without reverse transcriptase (RT + or −) was sequentially hybridized to each probe. Results for a representative experiment are shown; duplicate experiments were performed. (A) Asxl1 is expressed in unfractionated E14.5 fetal liver (TFL), and the hematopoietic stem cell–depleted (Sca-1−Lin+ [S−L+]), low (Sca-1+Lin+ [S+L+]), and enriched (Sca-1+Lin− [S+L−]) subpopulations, whereas Bmi1 expression is predominant in the 2 Sca-1+ fractions. (B) Asxl1 is expressed in unfractionated adult bone marrow (TBM) and in all fractionated adult bone marrow, spleen, and thymus cells investigated, whereas Bmi1 is expressed predominantly within the progenitor-enriched Sca-1+ fractions and the Gr-1 low fraction of total bone marrow, with very low or undetectable expression in other fractions.
Figure 3
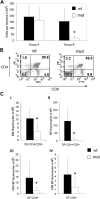
Asxl1tm1Bc/tm1Bc mutant mice exhibit reduced thymopoiesis. (A) Thymus cellularity is reduced in older (older than 15 weeks; group B, n = 8 each genotype) compared with younger (15 weeks or younger; group A, n = 6 wt, 4 mutant) Asxl1tm1Bc/tm1Bc (mut) adult mice compared with wild type. (B) Flow cytometry of T lymphocytes from adult thymus expressing the lineage markers CD4 and CD8 shows a relative increase in the double-negative and both single-positive fractions, and a relative decrease in the double-positive fraction in Asxl1tm1Bc/tm1Bc thymus compared with wild type. (C) Absolute cell number per thymus is reduced in group B Asxl1tm1Bc/tm1Bc mice compared with wild type for all fractions: (i) double-negative CD4−CD8− (DN), (ii) double-positive CD4+CD8+ (DP), (iii) single-positive (SP) CD4+, and (iv) single-positive CD8+ (n = 8). *P < .05.
Figure 4
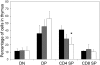
Flow cytometric analysis of T cells cultured from E14.5 thymus. Cells were analyzed using monoclonal antibodies against CD4 and CD8. Numbers indicate the percentage of cells within each subcompartment: double-negative CD4−CD8− (DN), double-positive CD4+CD8+ (DP), single-positive CD4 (CD4+SP), and single-positive CD8 (CD8+SP). *P < .05. ■ indicates wild type (n = 2); ▩, Asxl1+/tm1Bc (n = 6); and □, Asxl1tm1Bc/tm1Bc mutant (n = 7).
Figure 5
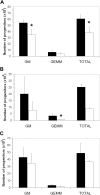
Asxl1tm1Bc/tm1Bc mutant mice exhibit reduced B-cell lymphopoiesis. (A) Flow cytometric profiles of adult (> 15 weeks old) peripheral blood (PB), spleen, and bone marrow (BM) show a relative decrease in B220- and IgM/IgD-positive cells in Asxl1tm1Bc/tm1Bc (mut) tissues compared with wild type (wt). (B) Absolute bone marrow cell numbers in adult femur expressing the markers B220; B220 and CD43; B220 and IgM; and IgM and IgD are significantly lower in Asxl1tm1Bc/tm1Bc mice compared with wild type (n = 8 each genotype except n = 4 for B220 and IgM). (C) Absolute cell numbers from > 15-week-old spleen expressing the markers B220, and IgM and IgD, are lower in Asxl1tm1Bc/tm1Bc mice compared with wild type (n = 8 each genotype). (D) In vitro colony formation of committed pre-B-lymphocyte progenitors (CFU–IL-7 assay) from adult bone marrow (> 15 weeks old) is significantly reduced in cultures of Asxl1tm1Bc/tm1Bc group B compared with wild type (n = 7 wt, 8 mutant). *P < .05.
Figure 6
Asxl1tm1Bc/tm1Bc mutant mice exhibit splenomegaly and increase in myeloid cell number. (A) Cellularity of Asxl1tm1Bc/tm1Bc (mut) adult spleens is increased compared with wild type (wt; n = 14 wt, 12 mutant). (B) In adult Asxl1tm1Bc/tm1Bc mice, the absolute numbers of myeloid (Gr1 or Mac1 positive) and erythroid (Ter119 positive) cells are increased in spleen (Gr1+ n = 13 wt, 11 mutant; Mac1+ n = 11; Ter119+ n = 10 wt, 7 mutant). (C) The proportion of cells bearing the myeloid lineage markers Gr1 (i) and Mac (ii) is increased in adult spleen of Asxl1tm1Bc/tm1Bc mutants.
Figure 7
Asxl1tm1Bc/tm1Bc mutant mice have fewer committed myeloerythroid progenitors. In vitro colony formation of committed myeloerythroid progenitors (CFU-GEMM assay) is reduced in Asxl1tm1Bc/tm1Bc mice compared with wild type in cultures of (A) E18.5 fetal liver (n = 2 wt, 3 mutant), (B) newborn spleen (n = 6 wt, 5 mutant), and (C) adult bone marrow (n = 9 wt, 6 mutant) *P < .05. ■ indicates wild type; □, Asxl1tm1Bc/tm1Bc mutant.
Similar articles
- The role of ASXL1 in hematopoiesis and myeloid malignancies.
Asada S, Fujino T, Goyama S, Kitamura T. Asada S, et al. Cell Mol Life Sci. 2019 Jul;76(13):2511-2523. doi: 10.1007/s00018-019-03084-7. Epub 2019 Mar 30. Cell Mol Life Sci. 2019. PMID: 30927018 Free PMC article. Review. - The distinct biological implications of Asxl1 mutation and its roles in leukemogenesis revealed by a knock-in mouse model.
Hsu YC, Chiu YC, Lin CC, Kuo YY, Hou HA, Tzeng YS, Kao CJ, Chuang PH, Tseng MH, Hsiao TH, Chou WC, Tien HF. Hsu YC, et al. J Hematol Oncol. 2017 Jul 11;10(1):139. doi: 10.1186/s13045-017-0508-x. J Hematol Oncol. 2017. PMID: 28697759 Free PMC article. - Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo.
Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM, Koche R, Shih AH, Guryanova OA, Kim E, Li S, Pandey S, Shin JY, Telis L, Liu J, Bhatt PK, Monette S, Zhao X, Mason CE, Park CY, Bernstein BE, Aifantis I, Levine RL. Abdel-Wahab O, et al. J Exp Med. 2013 Nov 18;210(12):2641-59. doi: 10.1084/jem.20131141. Epub 2013 Nov 11. J Exp Med. 2013. PMID: 24218140 Free PMC article. - Myelodysplastic syndromes are induced by histone methylation–altering ASXL1 mutations.
Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, Kagiyama Y, Kawabata KC, Nakahara F, Izawa K, Oki T, Maehara A, Isobe M, Tsuchiya A, Harada Y, Harada H, Ochiya T, Aburatani H, Kimura H, Thol F, Heuser M, Levine RL, Abdel-Wahab O, Kitamura T. Inoue D, et al. J Clin Invest. 2013 Nov;123(11):4627-40. doi: 10.1172/JCI70739. J Clin Invest. 2013. PMID: 24216483 Free PMC article. - Aberrant histone modifications induced by mutant ASXL1 in myeloid neoplasms.
Asada S, Kitamura T. Asada S, et al. Int J Hematol. 2019 Aug;110(2):179-186. doi: 10.1007/s12185-018-2563-7. Epub 2018 Dec 5. Int J Hematol. 2019. PMID: 30515738 Review.
Cited by
- Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes.
White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC; Sanger Institute Mouse Genetics Project; Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP. White JK, et al. Cell. 2013 Jul 18;154(2):452-64. doi: 10.1016/j.cell.2013.06.022. Cell. 2013. PMID: 23870131 Free PMC article. - Molecular pathophysiology of myelodysplastic syndromes.
Lindsley RC, Ebert BL. Lindsley RC, et al. Annu Rev Pathol. 2013 Jan 24;8:21-47. doi: 10.1146/annurev-pathol-011811-132436. Epub 2012 Aug 28. Annu Rev Pathol. 2013. PMID: 22934674 Free PMC article. Review. - The role of ASXL1 in hematopoiesis and myeloid malignancies.
Asada S, Fujino T, Goyama S, Kitamura T. Asada S, et al. Cell Mol Life Sci. 2019 Jul;76(13):2511-2523. doi: 10.1007/s00018-019-03084-7. Epub 2019 Mar 30. Cell Mol Life Sci. 2019. PMID: 30927018 Free PMC article. Review. - Disruption of asxl1 results in myeloproliferative neoplasms in zebrafish.
Gjini E, Jing CB, Nguyen AT, Reyon D, Gans E, Kesarsing M, Peterson J, Pozdnyakova O, Rodig SJ, Mansour MR, Joung K, Look AT. Gjini E, et al. Dis Model Mech. 2019 May 7;12(5):dmm035790. doi: 10.1242/dmm.035790. Dis Model Mech. 2019. PMID: 31064769 Free PMC article. - Disruption of the ASXL1 gene is frequent in primary, post-essential thrombocytosis and post-polycythemia vera myelofibrosis, but not essential thrombocytosis or polycythemia vera: analysis of molecular genetics and clinical phenotypes.
Stein BL, Williams DM, O'Keefe C, Rogers O, Ingersoll RG, Spivak JL, Verma A, Maciejewski JP, McDevitt MA, Moliterno AR. Stein BL, et al. Haematologica. 2011 Oct;96(10):1462-9. doi: 10.3324/haematol.2011.045591. Epub 2011 Jun 28. Haematologica. 2011. PMID: 21712540 Free PMC article.
References
- Brock HW, Fisher CL. Maintenance of gene expression patterns. Dev Dyn. 2005;232:633–655. - PubMed
- Brock HW, Lohuizen MV. The Polycomb group-no longer an exclusive club? Curr Opin Genet Dev. 2001;11:175–181. - PubMed
- Martinez AM, Schuettengruber B, Sakr S, Janic A, Gonzalez C, Cavalli G. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat Genet. 2009;41(10):1076–1082. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous