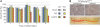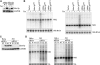Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance - PubMed (original) (raw)
Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance
Roman J Szczesny et al. Nucleic Acids Res. 2010 Jan.
Abstract
The mechanism of human mitochondrial RNA turnover and surveillance is still a matter of debate. We have obtained a cellular model for studying the role of hSuv3p helicase in human mitochondria. Expression of a dominant-negative mutant of the hSUV3 gene which encodes a protein with no ATPase or helicase activity results in perturbations of mtRNA metabolism and enables to study the processing and degradation intermediates which otherwise are difficult to detect because of their short half-lives. The hSuv3p activity was found to be necessary in the regulation of stability of mature, properly formed mRNAs and for removal of the noncoding processing intermediates transcribed from both H and L-strands, including mirror RNAs which represent antisense RNAs transcribed from the opposite DNA strand. Lack of hSuv3p function also resulted in accumulation of aberrant RNA species, molecules with extended poly(A) tails and degradation intermediates truncated predominantly at their 3'-ends. Moreover, we present data indicating that hSuv3p co-purifies with PNPase; this may suggest participation of both proteins in mtRNA metabolism.
Figures
Figure 1.
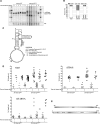
Expression and subcellular localization of hSuv3pWT and hSuv3pG207V in stable 293 cell lines. (A) Western blot analysis of hSuv3p level in 293 hSuv3pWT or 293 hSuv3pG207V cell lines. Expression of the exogenous genes was induced for 24 h with tetracycline at 25 ng/ml. GAPDH level is shown as the loading control. (B) Subcellular localization of endogenous hSuv3p (uninduced) and overproduced hSuv3pWT or hSuv3pG207V (induced) proteins. Exogenous gene expression was induced for 24 h and cells were subjected to immunofluorescence staining and confocal microscopy. The same microscope settings were used for uninduced and induced cells. Mitochondria were labeled with MitoTracker Red CMX Ros, hSuv3p with primary rabbit anti-hSuv3p antibodies followed by secondary AlexaFluor 488 conjugated antibodies. Nuclei were stained with Hoechst dye. The bar represents 10 μm.
Figure 2.
Effect of hSuv3pG207V expression on cell growth rate and cell morphology. (A) Graph representing growth rate of cells with integrated exogenous gene encoding wild-type (WT) or inactive form of hSuv3p (G207V). The growth rate of cells with empty vector integrated is also shown (Vector). Uninduced (–Tet) or induced (+Tet) cells were analyzed. The results represent the mean of three independent experiments. Error bars represent standard deviation. (B) Influence of hSuv3pG207V expression on cell morphology and color of the medium after 5 days of expression.
Figure 3.
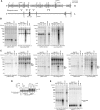
Expression of hSuv3pG207V affects mitochondrial RNA. Northern blot analysis of RNA isolated from uninduced or induced (+Tet) cells to overproduce wild-type or inactive form of hSuv3p. Impairment of hSuv3p function by expression of its inactive form leads to appearance of heterogeneous RNA (A and B) and affects (A) or not (B) the steady-state level of mRNA. Representative northern blots are shown. The graphs below show the levels of the analyzed mRNAs. Data represent the mean of three independent experiments, standard deviation is marked. The level of 7SL is shown as a loading control. (C) Northern blot analysis of ND3 mRNA in total RNA (Total), unbound (Unbound) and oligo-dT bound RNA fractions (Oligo-dT). A fragment of the membrane corresponding to the methylene blue staining of 18S rRNA is also shown for assessment of the fractionation efficiency.
Figure 4.
Influence of hSuv3pG207V expression on RNAs involved in mitochondrial translation. Changes in steady-state levels of 12S rRNA (A) and analyzed tRNAs (B). In both panels, representative northern blots are shown on the right. Graphs represent the mean of three independent experiments and standard deviation is marked.
Figure 5.
Expression of the non-mitochondrially localized hSuv3pWT or hSuv3pG207V does not affect mitochondrial RNA. (A) Western blot analysis of expression of the Δ46hSuv3pWT or Δ46hSuv3pG207V proteins. GAPDH level is shown as the loading control. (B) Northern blot analysis of ND2 and ND3 mRNAs. Cells were induced or not for 24 h, collected and RNA was isolated and analyzed by northern blot. Methylene blue staining of 18S rRNA is shown as loading control. (C and D) Silencing of hSUV3 expression by RNAi. HeLa and 293 cells were untreated (NT) or transfected with siRNAs noncomplementary to hSUV3 mRNA (N) or complementary to hSUV3 mRNA in three different places (S1, S2 and S3). (C) Western blot analysis of hSuv3p levels relative to GAPDH and (D) northern blot analysis of ND2 and ND3 mRNA levels relative to 7SL RNA.
Figure 6.
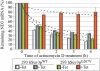
Impairment of hSuv3p function results in ND3 mRNA stabilization. Graph representing the level of ND3 mRNA measured by northern blot in cells untreated (point 0) and treated with the transcription inhibitor actinomycin D. Cells uninduced (–Tet) and induced (+Tet) for 24 h were analyzed. The results represent the mean of three independent experiments, error bars represent standard deviation, trend lines are also marked (dashed lines).
Figure 7.
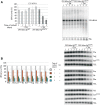
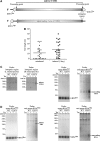
Effect of expression of hSuv3pG207V on transcript length. (A) High-resolution northern blot of ND3 mRNA; M, molecular weight marker. (B) Distribution of different classes of truncated clones identified by CRT-PCR. (C) Breaking points of 12S rRNA identified by CRT-PCR shown on a model of 12S rRNA structure. Boxed letters represent the first and the last nucleotide which can be analyzed by the applied assay. Numbers correspond to the nucleotide position in 12S rRNA (954 nt length). In some cases, the precise end of the truncated transcript cannot be identified as the adenine residue may belong to the poly(A) tail. See legend for details. (D) Analysis of the poly(A) tail length assessed by CRT-PCR. Each data point represents a single transcript. Polyadenylated transcripts [poly(A) tail longer than 25 nt] are shown as filled symbols and oligoadenylated ones (tail of 25 nt or less) as empty symbols. In cases where it was impossible to assess the precise length of the tail, because the adenine might belong to the tail or ‘body’ of the transcripts the average number was used. Horizontal lines represent average lengths of oligo- and polyadenylated fractions (E) Schematic representation of the two identified uridinylated transcripts.
Figure 8.
hSuv3p participates in removal of noncoding RNA. (A) Schematic representation of the polycistronic RNA resulting from H- or L-strand transcription and localization of the used riboprobes. The letters on the right refer to the template strand. (B–E) Northern blot analysis using strand-specific riboprobes. RNA isolated from uninduced cells or cells induced for 24 hours was used. (D) Northern blot analysis of the ND2 mirror transcript level in cells untreated (point 0) and treated with transcription inhibitor actinomycin D. Expression of the hSuv3pG207V had been induced for 24 h before actinomycin D was added. Graph on the top shows quantification of the signal by using ImageQuant software.
Figure 9.
Mapping the ends of mirror transcripts. (A) Schematic map of the COX1 region of mtDNA. Boxed arrows represent coding sequences. Processing points identified by CRT-PCR are marked. (B) Analysis of the poly(A) tail length of the COX1 mirror transcript assessed by CRT-PCR. Symbols represent single transcripts. RNA isolated from 293 hSuv3pG207V cells uninduced and induced for 24 h was used. Horizontal lines represent average lengths of oligo- and polyadenylated fractions (C) Northern blot analysis confirming the CRT-PCR analysis of the COX1 mirror transcript. Strand-specific probes were used. (D) Northern blot analysis of the 3′-end of the ND2 mirror transcript. Strand-specific probes were used. See text for details and Figures 8A and 9A for a schematic representation of the polycistronic transcripts.
Figure 10.
Co-purification of hSuv3p with PNPase. The mtTAP, hSuv3TAP and PNPTAP correspond to the name of the cell line and the type of expressed TAP fusion protein. The mtTAP is a short polypeptide which probably causes its low stability and lower expression than hSuv3TAP. (A and B) Western blot analysis of PNPase presence in total (Total), mitochondrial (Mito) lysates and elution (Elution) fraction obtained after one- (A) or two-step (B) chromatography. The mtTAP and hSuv3TAP fusion proteins were detected because the secondary antibodies used for PNPase detection bind directly to the A protein, a TAP tag component. (C) Western blot analysis of the hSuv3p presence in total (Total), mitochondrial (Mito) lysates and elution (Elution) fraction. The PNPTAP fusion protein was detected because the rabbit primary anti-hSuv3p antibodies bind directly to the A protein, a TAP tag component. (D) Western blot analysis of hSuv3p and PNPase presence in fractions obtained by size exclusion chromatography. The migration of the marker proteins is shown. Fractions in which hSuv3p and PNPase migrate together are boxed.
Similar articles
- Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci.
Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Borowski LS, et al. Nucleic Acids Res. 2013 Jan;41(2):1223-40. doi: 10.1093/nar/gks1130. Epub 2012 Dec 5. Nucleic Acids Res. 2013. PMID: 23221631 Free PMC article. - RNA degradation in human mitochondria: the journey is not finished.
Santonoceto G, Jurkiewicz A, Szczesny RJ. Santonoceto G, et al. Hum Mol Genet. 2024 May 22;33(R1):R26-R33. doi: 10.1093/hmg/ddae043. Hum Mol Genet. 2024. PMID: 38779774 Free PMC article. Review. - SUV3 helicase is required for correct processing of mitochondrial transcripts.
Clemente P, Pajak A, Laine I, Wibom R, Wedell A, Freyer C, Wredenberg A. Clemente P, et al. Nucleic Acids Res. 2015 Sep 3;43(15):7398-413. doi: 10.1093/nar/gkv692. Epub 2015 Jul 7. Nucleic Acids Res. 2015. PMID: 26152302 Free PMC article. - LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria.
Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T. Chujo T, et al. Nucleic Acids Res. 2012 Sep;40(16):8033-47. doi: 10.1093/nar/gks506. Epub 2012 May 31. Nucleic Acids Res. 2012. PMID: 22661577 Free PMC article. - RNA turnover in human mitochondria: more questions than answers?
Borowski LS, Szczesny RJ, Brzezniak LK, Stepien PP. Borowski LS, et al. Biochim Biophys Acta. 2010 Jun-Jul;1797(6-7):1066-70. doi: 10.1016/j.bbabio.2010.01.028. Epub 2010 Feb 2. Biochim Biophys Acta. 2010. PMID: 20117077 Review.
Cited by
- Human PNPase causes RNA stabilization and accumulation of R-loops in the Escherichia coli model system.
Falchi FA, Forti F, Carnelli C, Genco A, Pizzoccheri R, Manzari C, Pavesi G, Briani F. Falchi FA, et al. Sci Rep. 2023 Jul 21;13(1):11771. doi: 10.1038/s41598-023-38924-x. Sci Rep. 2023. PMID: 37479726 Free PMC article. - Spatial analysis of mitochondrial gene expression reveals dynamic translation hubs and remodeling in stress.
Begeman A, Smolka JA, Shami A, Waingankar TP, Lewis SC. Begeman A, et al. Sci Adv. 2025 Apr 18;11(16):eads6830. doi: 10.1126/sciadv.ads6830. Epub 2025 Apr 18. Sci Adv. 2025. PMID: 40249810 Free PMC article. - The human Suv3 helicase interacts with replication protein A and flap endonuclease 1 in the nucleus.
Venø ST, Kulikowicz T, Pestana C, Stepien PP, Stevnsner T, Bohr VA. Venø ST, et al. Biochem J. 2011 Dec 1;440(2):293-300. doi: 10.1042/BJ20100991. Biochem J. 2011. PMID: 21846330 Free PMC article. - Mitochondrial Polyadenylation Is a One-Step Process Required for mRNA Integrity and tRNA Maturation.
Bratic A, Clemente P, Calvo-Garrido J, Maffezzini C, Felser A, Wibom R, Wedell A, Freyer C, Wredenberg A. Bratic A, et al. PLoS Genet. 2016 May 13;12(5):e1006028. doi: 10.1371/journal.pgen.1006028. eCollection 2016 May. PLoS Genet. 2016. PMID: 27176048 Free PMC article. - Overexpression of PDH45 or SUV3 helicases in rice leads to delayed leaf senescence-associated events.
Macovei A, Sahoo RK, Faè M, Balestrazzi A, Carbonera D, Tuteja N. Macovei A, et al. Protoplasma. 2017 Mar;254(2):1103-1113. doi: 10.1007/s00709-016-1017-4. Epub 2016 Sep 1. Protoplasma. 2017. PMID: 27586643
References
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. - PubMed
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. - PubMed
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. - PubMed
- Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases