Predicting new molecular targets for known drugs - PubMed (original) (raw)
. 2009 Nov 12;462(7270):175-81.
doi: 10.1038/nature08506. Epub 2009 Nov 1.
Vincent Setola, John J Irwin, Christian Laggner, Atheir I Abbas, Sandra J Hufeisen, Niels H Jensen, Michael B Kuijer, Roberto C Matos, Thuy B Tran, Ryan Whaley, Richard A Glennon, Jérôme Hert, Kelan L H Thomas, Douglas D Edwards, Brian K Shoichet, Bryan L Roth
Affiliations
- PMID: 19881490
- PMCID: PMC2784146
- DOI: 10.1038/nature08506
Predicting new molecular targets for known drugs
Michael J Keiser et al. Nature. 2009.
Abstract
Although drugs are intended to be selective, at least some bind to several physiological targets, explaining side effects and efficacy. Because many drug-target combinations exist, it would be useful to explore possible interactions computationally. Here we compared 3,665 US Food and Drug Administration (FDA)-approved and investigational drugs against hundreds of targets, defining each target by its ligands. Chemical similarities between drugs and ligand sets predicted thousands of unanticipated associations. Thirty were tested experimentally, including the antagonism of the beta(1) receptor by the transporter inhibitor Prozac, the inhibition of the 5-hydroxytryptamine (5-HT) transporter by the ion channel drug Vadilex, and antagonism of the histamine H(4) receptor by the enzyme inhibitor Rescriptor. Overall, 23 new drug-target associations were confirmed, five of which were potent (<100 nM). The physiological relevance of one, the drug N,N-dimethyltryptamine (DMT) on serotonergic receptors, was confirmed in a knockout mouse. The chemical similarity approach is systematic and comprehensive, and may suggest side-effects and new indications for many drugs.
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
Figures
Figure 1. Drug-target networks, before and after predicting off-targets
(A) Known drug-target network. Each drug (gold) is linked to its known protein targets (cyan) by a gray edge. Each edge denotes a _K_i of 1 μM or better for that drug to its target. (B) Predicted drug-target network. Drugs and proteins are linked as per the known drug-target network in (A), but with the addition of red edges representing SEA off-target predictions with E-values ≤ 10-10.
Figure 2. Testing new off-target activities
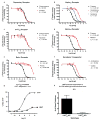
(A-F) Radioligand competition binding assays: (A) Doralese at D4, (B) Sedalande and Dimetholizine at α1D, (C) Fabahistin at 5-HT5A, (D) Motilium at α1A, (E) Prozac at β1, and (F) Vadilex at the serotonin transporter. (G-H) Investigating 5-HT2A as the target of DMT-induced hallucination: (G) 5-HT2A-mediated Ca2+ response was measured after treating HEK 293 cells stably expressing the human 5-HT2A receptor with DMT or 5-HT. DMT's EC50 was found to be 118±29 nM (vs. 5-HT's 6.6±0.4 nM baseline, n = 3), with an Emax of 23±0.4% (n = 3), confirming that DMT is a potent partial agonist at 5-HT2A receptors. (H) DMT elicited head twitch behavior only in 5-HT2A wild-type mice, confirming that it is a hallucinogenic 5-HT2A agonist. **, p < .01.
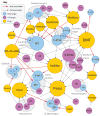
Figure 3. Discovered off-targets network
Bipartite network where drugs (gold) are linked by gray edges to their known targets (violet) and by red arrows to their discovered off-targets (cyan). Gray edges denote binding at 1 μM or better, where these affinities are known. Node sizes increase with number of incident edges. Target abbreviations: 5-HT_x_, serotonin receptor type x; 5-HTT, serotonin transporter; β1+, β1 adrenergic agonist; β1-, β1 adrenergic antagonist; β3+, β3 adrenergic agonist; σ1, σ1-receptor; CA, carbonic anhydrase; DAT, dopamine transporter; HIV1RT, HIV-1 reverse transcriptase; hERG, human Ether-a-go-go Related Gene channel; K+, Potassium channel; NET, norepinephrine transporter; NMDA, _N_-methyl-_D_-aspartate receptor; VMAT2, vesicular monoamine transporter 2.
Comment in
- Drug discovery: Predicting promiscuity.
Hopkins AL. Hopkins AL. Nature. 2009 Nov 12;462(7270):167-8. doi: 10.1038/462167a. Nature. 2009. PMID: 19907483 No abstract available. - Antabuse (disulfiram) as a pilot case of nonprofit drug.
Cvek B. Cvek B. Int J Cancer. 2010 Nov 15;127(10):2486. doi: 10.1002/ijc.25237. Int J Cancer. 2010. PMID: 20143396 No abstract available.
Similar articles
- Predicting new indications for approved drugs using a proteochemometric method.
Dakshanamurthy S, Issa NT, Assefnia S, Seshasayee A, Peters OJ, Madhavan S, Uren A, Brown ML, Byers SW. Dakshanamurthy S, et al. J Med Chem. 2012 Aug 9;55(15):6832-48. doi: 10.1021/jm300576q. Epub 2012 Jul 25. J Med Chem. 2012. PMID: 22780961 Free PMC article. - Drug target identification using side-effect similarity.
Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Campillos M, et al. Science. 2008 Jul 11;321(5886):263-6. doi: 10.1126/science.1158140. Science. 2008. PMID: 18621671 - Drug discovery: Predicting promiscuity.
Hopkins AL. Hopkins AL. Nature. 2009 Nov 12;462(7270):167-8. doi: 10.1038/462167a. Nature. 2009. PMID: 19907483 No abstract available. - The chemical basis of pharmacology.
Keiser MJ, Irwin JJ, Shoichet BK. Keiser MJ, et al. Biochemistry. 2010 Dec 7;49(48):10267-76. doi: 10.1021/bi101540g. Epub 2010 Nov 12. Biochemistry. 2010. PMID: 21058655 Free PMC article. Review. - Preclinical pharmacokinetics: an approach towards safer and efficacious drugs.
Singh SS. Singh SS. Curr Drug Metab. 2006 Feb;7(2):165-82. doi: 10.2174/138920006775541552. Curr Drug Metab. 2006. PMID: 16472106 Review.
Cited by
- A novel multi-modal drug repurposing approach for identification of potent ACK1 inhibitors.
Phatak SS, Zhang S. Phatak SS, et al. Pac Symp Biocomput. 2013:29-40. Pac Symp Biocomput. 2013. PMID: 23424109 Free PMC article. - Introducing the metacore concept for multi-target ligand design.
Stumpfe D, Hoch A, Bajorath J. Stumpfe D, et al. RSC Med Chem. 2021 Apr 15;12(4):628-635. doi: 10.1039/d1md00056j. eCollection 2021 Apr 28. RSC Med Chem. 2021. PMID: 34046634 Free PMC article. - Frentizole, a Nontoxic Immunosuppressive Drug, and Its Analogs Display Antitumor Activity via Tubulin Inhibition.
Ramos S, Vicente-Blázquez A, López-Rubio M, Gallego-Yerga L, Álvarez R, Peláez R. Ramos S, et al. Int J Mol Sci. 2023 Dec 14;24(24):17474. doi: 10.3390/ijms242417474. Int J Mol Sci. 2023. PMID: 38139302 Free PMC article. - The pharmacological interaction of compounds in ayahuasca: a systematic review.
Ruffell S, Netzband N, Bird C, Young AH, Juruena MF. Ruffell S, et al. Braz J Psychiatry. 2020 Nov-Dec;42(6):646-656. doi: 10.1590/1516-4446-2020-0884. Braz J Psychiatry. 2020. PMID: 32638916 Free PMC article. - Hybrid phenotype mining method for investigating off-target protein and underlying side effects of anti-tumor immunotherapy.
Zheng Y, Meng X, Zweigenbaum P, Chen L, Xia J. Zheng Y, et al. BMC Med Inform Decis Mak. 2020 Jul 9;20(Suppl 3):133. doi: 10.1186/s12911-020-1105-4. BMC Med Inform Decis Mak. 2020. PMID: 32646421 Free PMC article.
References
- Ehrlich P. The Theory and Practice of Chemotherapy. Folia Serologica. 1911;7:697–714.
- Peterson RT. Chemical biology and the limits of reductionism. Nat Chem Biol. 2008;4:635–638. - PubMed
- Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27:157–167. - PubMed
- Marona-Lewicka D, Nichols DE. Further evidence that the delayed temporal dopaminergic effects of LSD are mediated by a mechanism different than the first temporal phase of action. Pharmacol Biochem Behav. 2007;87:453–461. - PubMed
- Marona-Lewicka D, Nichols DE. WAY 100635 produces discriminative stimulus effects in rats mediated by dopamine D(4) receptor activation. Behav Pharmacol. 2009;20:114–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 MH082441-029002/MH/NIMH NIH HHS/United States
- U19 MH082441/MH/NIMH NIH HHS/United States
- R01 MH061887-09/MH/NIMH NIH HHS/United States
- U19 MH082441-019003/MH/NIMH NIH HHS/United States
- U19 MH082441-03/MH/NIMH NIH HHS/United States
- U19 MH082441-039002/MH/NIMH NIH HHS/United States
- U19 MH082441-030001/MH/NIMH NIH HHS/United States
- U19 MH082441-010001/MH/NIMH NIH HHS/United States
- U19 MH082441-02/MH/NIMH NIH HHS/United States
- R01 MH061887/MH/NIMH NIH HHS/United States
- R01 DA017204-04/DA/NIDA NIH HHS/United States
- R01 DA017204-05/DA/NIDA NIH HHS/United States
- U19 MH082441-01/MH/NIMH NIH HHS/United States
- U19 MH082441-020001/MH/NIMH NIH HHS/United States
- U19 MH082441-019002/MH/NIMH NIH HHS/United States
- R01 DA017204/DA/NIDA NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R01 MH061887-10/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical