Microdroplet-based PCR enrichment for large-scale targeted sequencing - PubMed (original) (raw)
. 2009 Nov;27(11):1025-31.
doi: 10.1038/nbt.1583. Epub 2009 Nov 1.
Jason B Warner, Masakazu Nakano, Brian Libby, Martina Medkova, Patricia H David, Steve K Kotsopoulos, Michael L Samuels, J Brian Hutchison, Jonathan W Larson, Eric J Topol, Michael P Weiner, Olivier Harismendy, Jeff Olson, Darren R Link, Kelly A Frazer
Affiliations
- PMID: 19881494
- PMCID: PMC2779736
- DOI: 10.1038/nbt.1583
Microdroplet-based PCR enrichment for large-scale targeted sequencing
Ryan Tewhey et al. Nat Biotechnol. 2009 Nov.
Erratum in
- Nat Biotechnol. 2010 Feb;28(2):178
Abstract
Targeted enrichment of specific loci of the human genome is a promising approach to enable sequencing-based studies of genetic variation in large populations. Here we describe an enrichment approach based on microdroplet PCR, which enables 1.5 million amplifications in parallel. We sequenced six samples enriched by microdroplet or traditional singleplex PCR using primers targeting 435 exons of 47 genes. Both methods generated similarly high-quality data: 84% of the uniquely mapping reads fell within the targeted sequences; coverage was uniform across approximately 90% of targeted bases; sequence variants were called with >99% accuracy; and reproducibility between samples was high (r(2) = 0.9). We scaled the microdroplet PCR to 3,976 amplicons totaling 1.49 Mb of sequence, sequenced the resulting sample with both Illumina GAII and Roche 454, and obtained data with equally high specificity and sensitivity. Our results demonstrate that microdroplet technology is well suited for processing DNA for massively parallel enrichment of specific subsets of the human genome for targeted sequencing.
Figures
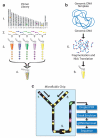
Figure 1. Microdroplet PCR workflow
Primer Library Generation (A): (1) Identify targeted sequences of interest in the genome. (2) Design and synthesize forward and reverse primer pairs for each targeted sequence (library element). (3) Generation of primer pair droplets for each library element. A microfluidic chip is used to encapsulate the aqueous PCR primers in inert fluorinated carrier oil with a block-copolymer surfactant to generate the equivalent of a picoliter scale test tube compatible with standard molecular biology. (4) Primer library, primer pair droplets of library elements are mixed together so that each library element has an equal representation. Genomic DNA Template Mix Preparation (B): (5) Genomic DNA is biotinylated (red dots), fragmented into 2 to 4 kb fragments and purified. (6) Purified genomic DNA is mixed together with all of the components of the PCR reaction (DNA polymerase, dNTPs, and buffer) except for the PCR primers. Droplet Merge and PCR (C): (7) Primer Library droplets are dispensed to the microfluidic chip (8) while the Genomic DNA Template is delivered as an aqueous solution and template droplets are formed within the microfluidic chip. The primer pair droplets and template droplets are then paired together in a 1:1 ratio. (9) Paired droplets flow through the channel of the microfluidic chip to pass through a merge area where an electric field induces the two discrete droplets to coalesce into a single PCR droplet. The roughly 1.5 million PCR droplets are collected into a single 0.2 ml PCR tube. The collection of PCR droplets (PCR Library) is processed in a standard thermal cycler for targeted amplification, followed by breaking the emulsion of PCR droplets to release the PCR amplicons into solution for genomic DNA (gDNA) removal, purification and sequencing.
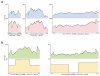
Figure 2. Coverage plots of targeted sequences
For the validation phase (A) base by base coverage of three target sequences selected for their varying lengths and GC% amplified by microdroplet (blue) and traditional (red) PCR. For the scale-up phase (B) the coverage of two targets representing an average and maximum amplicon length sequenced by Illumina GA (green) and Roche 454 (yellow) is shown. At the bottom of each plot the PCR primer positions (grey dumbbells connected by line) are shown. Roche 454 end sequencing of average sized amplicons results in 2-fold higher coverage of middle bases whereas end sequencing of larger amplicons results in middle bases having no coverage.
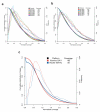
Figure 3. Normalized Coverage Distribution Plots
The validation phase 457 amplicons amplified by traditional PCR (A) and microdroplet PCR (B) and the scale-up phase 3976 amplicons amplified by microdroplet PCR (C). Normalized coverage is the absolute base coverage divided by the mean coverage of bases for the indicated sample. Each colored line represents either one of the six samples (A & B) or one of two sequencing platforms (C). The solid colored lines represent the cumulative distribution (left axis) for each sample. The colored dashed lines indicate a skewed normal distribution (right axis) for each sample. For each sample the mean coverage across all bases are listed.
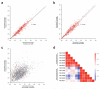
Figure 4. Inter-sample reproducibility of amplicon coverage
For the validation phase the normalized mean coverage of each amplicon is plotted for NA12006 (Caucasian) versus NA18505 (African) samples for the traditional (A) and microdroplet (B) PCR methods. For each sample (assigned same color as in Figure 2) the average normalized coverage of each amplicon is plotted for traditional versus microdroplet (C). Correlation matrix for all samples depicting Lin's concordance coefficient (D). All samples show a high correlation among each other within a PCR method but not between the two methods.
Comment in
- Targeted sequencing with microfluidics.
Kirkness EF. Kirkness EF. Nat Biotechnol. 2009 Nov;27(11):998-9. doi: 10.1038/nbt1109-998. Nat Biotechnol. 2009. PMID: 19898452 No abstract available.
Similar articles
- Mung bean nuclease treatment increases capture specificity of microdroplet-PCR based targeted DNA enrichment.
Yu Z, Cao K, Tischler T, Stolle CA, Santani AB. Yu Z, et al. PLoS One. 2014 Jul 24;9(7):e103491. doi: 10.1371/journal.pone.0103491. eCollection 2014. PLoS One. 2014. PMID: 25058678 Free PMC article. - Assessment of target enrichment platforms using massively parallel sequencing for the mutation detection for congenital muscular dystrophy.
Valencia CA, Rhodenizer D, Bhide S, Chin E, Littlejohn MR, Keong LM, Rutkowski A, Bonnemann C, Hegde M. Valencia CA, et al. J Mol Diagn. 2012 May-Jun;14(3):233-46. doi: 10.1016/j.jmoldx.2012.01.009. Epub 2012 Mar 16. J Mol Diagn. 2012. PMID: 22426012 Free PMC article. - Microdroplet PCR for Highly Multiplexed Targeted Bisulfite Sequencing.
Komori HK, LaMere SA, Hart T, Head SR, Torkamani A, Salomon DR. Komori HK, et al. Methods Mol Biol. 2018;1708:333-348. doi: 10.1007/978-1-4939-7481-8_17. Methods Mol Biol. 2018. PMID: 29224152 - Massively parallel sequencing of ataxia genes after array-based enrichment.
Hoischen A, Gilissen C, Arts P, Wieskamp N, van der Vliet W, Vermeer S, Steehouwer M, de Vries P, Meijer R, Seiqueros J, Knoers NV, Buckley MF, Scheffer H, Veltman JA. Hoischen A, et al. Hum Mutat. 2010 Apr;31(4):494-9. doi: 10.1002/humu.21221. Hum Mutat. 2010. PMID: 20151403 - Performance evaluation of the next-generation sequencing approach for molecular diagnosis of hereditary hearing loss.
Sivakumaran TA, Husami A, Kissell D, Zhang W, Keddache M, Black AP, Tinkle BT, Greinwald JH Jr, Zhang K. Sivakumaran TA, et al. Otolaryngol Head Neck Surg. 2013 Jun;148(6):1007-16. doi: 10.1177/0194599813482294. Epub 2013 Mar 22. Otolaryngol Head Neck Surg. 2013. PMID: 23525850
Cited by
- Genotyping single nucleotide polymorphisms in homologous regions using multiplex kb level amplicon capture sequencing.
Lu M, Li J, Sun X, Zhao D, Zong H, Tang C, Li K, Zhou Y, Xiao J. Lu M, et al. Mol Genet Genomics. 2024 Oct 26;299(1):99. doi: 10.1007/s00438-024-02192-9. Mol Genet Genomics. 2024. PMID: 39460824 - A Review of Probe-Based Enrichment Methods to Inform Plant Virus Diagnostics.
Farrall T, Brawner J, Dinsdale A, Kehoe M. Farrall T, et al. Int J Mol Sci. 2024 Jul 30;25(15):8348. doi: 10.3390/ijms25158348. Int J Mol Sci. 2024. PMID: 39125919 Free PMC article. Review. - Dielectrophoretic bead-droplet reactor for solid-phase synthesis.
Padhy P, Zaman MA, Jensen MA, Cheng YT, Huang Y, Wu M, Galambos L, Davis RW, Hesselink L. Padhy P, et al. Nat Commun. 2024 Jul 22;15(1):6159. doi: 10.1038/s41467-024-49284-z. Nat Commun. 2024. PMID: 39039069 Free PMC article. - Fast Thermocycling in Custom Microfluidic Cartridge for Rapid Single-Molecule Droplet PCR.
Takahara H, Tanaka H, Hashimoto M. Takahara H, et al. Sensors (Basel). 2023 Dec 17;23(24):9884. doi: 10.3390/s23249884. Sensors (Basel). 2023. PMID: 38139729 Free PMC article. - Robust and rapid partitioning in thermoplastic.
Quan PL, Alvarez-Amador M, Jiang Y, Sauzade M, Brouzes E. Quan PL, et al. Analyst. 2023 Dec 18;149(1):100-107. doi: 10.1039/d3an01869e. Analyst. 2023. PMID: 37982399
References
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. - PubMed
Publication types
MeSH terms
Grants and funding
- 1R21CA125693-01/CA/NCI NIH HHS/United States
- UL1 RR025774/RR/NCRR NIH HHS/United States
- 1U54RR025204-01/RR/NCRR NIH HHS/United States
- P50 HL077107/HL/NHLBI NIH HHS/United States
- P50 HL077107-020001/HL/NHLBI NIH HHS/United States
- R21 CA125693/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous