Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer - PubMed (original) (raw)
Review
. 2010 Jan 21;29(3):313-24.
doi: 10.1038/onc.2009.358. Epub 2009 Nov 2.
Affiliations
- PMID: 19881548
- PMCID: PMC2809806
- DOI: 10.1038/onc.2009.358
Review
Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer
R J DeBerardinis et al. Oncogene. 2010.
Abstract
Several decades of research have sought to characterize tumor cell metabolism in the hope that tumor-specific activities can be exploited to treat cancer. Having originated from Warburg's seminal observation of aerobic glycolysis in tumor cells, most of this attention has focused on glucose metabolism. However, since the 1950s cancer biologists have also recognized the importance of glutamine (Q) as a tumor nutrient. Glutamine contributes to essentially every core metabolic task of proliferating tumor cells: it participates in bioenergetics, supports cell defenses against oxidative stress and complements glucose metabolism in the production of macromolecules. The interest in glutamine metabolism has been heightened further by the recent findings that c-myc controls glutamine uptake and degradation, and that glutamine itself exerts influence over a number of signaling pathways that contribute to tumor growth. These observations are stimulating a renewed effort to understand the regulation of glutamine metabolism in tumors and to develop strategies to target glutamine metabolism in cancer. In this study we review the protean roles of glutamine in cancer, both in the direct support of tumor growth and in mediating some of the complex effects on whole-body metabolism that are characteristic of tumor progression.
Figures
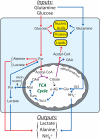
Figure 1. Glutamine supports cell survival, growth and proliferation through metabolic and non-metabolic mechanisms
After its import through surface transporters like SLC1A5, glutamine (Gln) is either exported in exchange with the import of essential amino acids (EAA) or consumed in various pathways that together support the basic metabolic functions needed for cell survival, growth and proliferation. In cancer cells, the mitochondrial enzyme glutaminase (GLS) appears to account for the largest fraction of net glutamine consumption. This enzyme produces NH4+, which is exported, perhaps through carrier-mediated mechanisms. Abbreviations: mTORC1, mammalian Target of Rapamycin Complex 1; Glu, glutamate; NEAA, nonessential amino acids; Cys, cysteine; Cys-Cys, cystine; GSH, glutathione; mGluR, metabotropic glutamate receptor; ERK, extracellular signal-regulated protein kinase; PI3K, phosphatidylinositol 3’-kinase; α-KG, α-ketoglutarate; Mal, malate; OAA, oxaloacetate; Ac-CoA, acetyl-CoA; Cit, citrate; Pyr, pyruvate; Lac, lactate; TCA, tricarboxylic acid; GLS, glutaminase; ME, malic enzyme.
Figure 2. Cooperativity between glucose and glutamine metabolism in growing tumors
The major nutrients consumed by tumors are glutamine and glucose, which provide precursors for nucleic acids, proteins and lipids, the three classes of macromolecules needed to produce daughter cells. The metabolism of glutamine (blue arrows) and glucose (red arrows) are complementary, converging on the production of citrate (purple arrows). Glutamine metabolism produces oxaloacetate (OAA) and NADPH, both of which are required to convert glucose carbon into macromolecules. Glutamine metabolism also supplements the pyruvate pool, which is predominantly formed from glucose. As a consequence of the rapid metabolism of these two nutrients, lactate, alanine and NH4+ are secreted by the tumor. Abbreviations: GLS, glutaminase; Glu, glutamate; α-KG, α-ketoglutarate; Succ, succinate; Fum, fumarate; Mal, malate; Pyr, pyruvate; Lac, lactate; TCA, tricarboxylic acid.
Figure 3. Proposed inter-organ metabolic cycles in cachectic cancer patients
As summarized in Figure 2, growth of the tumor involves consumption of glucose and glutamine with secretion of lactate, alanine and ammonia. Some of the lactate may be taken up by well-oxygenated regions of the tumor and used as a respiratory fuel. Other lactate and alanine are delivered to the liver and used to produce glucose, which can then return to the tumor (the Cori cycle). Meanwhile, the ammonia can be disposed through the urea cycle, or possibly delivered to the muscle for incorporation into new glutamine molecules produced during protein catabolism and glucose metabolism. Both the Cori cycle and the putative glutamine-ammonia cycle deliver energy to the tumor, but cost energy in the other organs involved, driving up whole-body energy expenditure as is typically observed in cancer cachexia. Glu, glutamate; α-KG, α-ketoglutarate; AAs, amino acids; GLUL, glutamate-ammonia ligase (i.e. glutamine synthetase).
Similar articles
- Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis.
Daye D, Wellen KE. Daye D, et al. Semin Cell Dev Biol. 2012 Jun;23(4):362-9. doi: 10.1016/j.semcdb.2012.02.002. Epub 2012 Feb 11. Semin Cell Dev Biol. 2012. PMID: 22349059 Review. - Rethinking the Warburg effect with Myc micromanaging glutamine metabolism.
Dang CV. Dang CV. Cancer Res. 2010 Feb 1;70(3):859-62. doi: 10.1158/0008-5472.CAN-09-3556. Epub 2010 Jan 19. Cancer Res. 2010. PMID: 20086171 Free PMC article. Review. - Multi-scale computational study of the Warburg effect, reverse Warburg effect and glutamine addiction in solid tumors.
Shan M, Dai D, Vudem A, Varner JD, Stroock AD. Shan M, et al. PLoS Comput Biol. 2018 Dec 7;14(12):e1006584. doi: 10.1371/journal.pcbi.1006584. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30532226 Free PMC article. - Glutamine at focus: versatile roles in cancer.
De Vitto H, Pérez-Valencia J, Radosevich JA. De Vitto H, et al. Tumour Biol. 2016 Feb;37(2):1541-58. doi: 10.1007/s13277-015-4671-9. Epub 2015 Dec 24. Tumour Biol. 2016. PMID: 26700676 Review. - Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells.
Chen JQ, Russo J. Chen JQ, et al. Biochim Biophys Acta. 2012 Dec;1826(2):370-84. doi: 10.1016/j.bbcan.2012.06.004. Epub 2012 Jun 27. Biochim Biophys Acta. 2012. PMID: 22750268 Review.
Cited by
- New insights into the roles of lactylation in cancer.
Zhu Y, Liu W, Luo Z, Xiao F, Sun B. Zhu Y, et al. Front Pharmacol. 2024 Oct 22;15:1412672. doi: 10.3389/fphar.2024.1412672. eCollection 2024. Front Pharmacol. 2024. PMID: 39502530 Free PMC article. Review. - Animal Models of Retinopathy of Prematurity: Advances and Metabolic Regulators.
Maurya M, Liu CH, Bora K, Kushwah N, Pavlovich MC, Wang Z, Chen J. Maurya M, et al. Biomedicines. 2024 Aug 23;12(9):1937. doi: 10.3390/biomedicines12091937. Biomedicines. 2024. PMID: 39335451 Free PMC article. Review. - Glutamine promotes the proliferation of intestinal stem cells via inhibition of TP53-induced glycolysis and apoptosis regulator promoter methylation in burned mice.
Zhang P, Wu D, Zha X, Su S, Zhang Y, Wei Y, Xia L, Fan S, Peng X. Zhang P, et al. Burns Trauma. 2024 Sep 26;12:tkae045. doi: 10.1093/burnst/tkae045. eCollection 2024. Burns Trauma. 2024. PMID: 39328365 Free PMC article. - KDM4A promotes malignant progression of breast cancer by down-regulating BMP9 inducing consequent enhancement of glutamine metabolism.
Chen Y, Yang S, Yu T, Zeng T, Wei L, You Y, Tang J, Dang T, Sun H, Zhang Y. Chen Y, et al. Cancer Cell Int. 2024 Sep 19;24(1):322. doi: 10.1186/s12935-024-03504-0. Cancer Cell Int. 2024. PMID: 39300582 Free PMC article. - Glutamine Metabolism and Prostate Cancer.
Erb HHH, Polishchuk N, Stasyk O, Kahya U, Weigel MM, Dubrovska A. Erb HHH, et al. Cancers (Basel). 2024 Aug 18;16(16):2871. doi: 10.3390/cancers16162871. Cancers (Basel). 2024. PMID: 39199642 Free PMC article. Review.
References
- Abcouwer SF, Schwarz C, Meguid RA. Glutamine deprivation induces the expression of GADD45 and GADD153 primarily by mRNA stabilization. J Biol Chem. 1999;274:28645–51. - PubMed
- Bangsbo J, Kiens B, Richter EA. Ammonia uptake in inactive muscles during exercise in humans. Am J Physiol. 1996;270:E101–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources