Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells - PubMed (original) (raw)
. 2009 Dec;119(12):3765-73.
doi: 10.1172/JCI40732. Epub 2009 Nov 2.
Mickaël M Ménager, Agathe Burgess, Nizar Mahlaoui, Capucine Picard, Catherine Schaffner, Fahad Al-Manjomi, Musa Al-Harbi, Abdullah Alangari, Françoise Le Deist, Andrew R Gennery, Nathalie Prince, Astrid Cariou, Patrick Nitschke, Ulrich Blank, Gehad El-Ghazali, Gaël Ménasché, Sylvain Latour, Alain Fischer, Geneviève de Saint Basile
Affiliations
- PMID: 19884660
- PMCID: PMC2786810
- DOI: 10.1172/JCI40732
Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells
Marjorie Côte et al. J Clin Invest. 2009 Dec.
Abstract
Familial hemophagocytic lymphohistiocytosis (FHL) is a genetically heterogeneous autosomal recessive immune disorder characterized by the occurrence of uncontrolled activation of lymphocytes and macrophages infiltrating multiple organs. Disease-causing mutations in the perforin (PRF1; also known as FHL2), Munc13-4 (UNC13D; also known as FHL3), and syntaxin-11 (STX11; also known as FHL4) genes have been identified in individuals with FHL. These genes all encode proteins involved in the cytotoxic activity of lymphocytes. Here, we show that the gene encoding syntaxin-binding protein 2 (Munc18-2; official gene symbol STXBP2) is mutated in another subset of patients with FHL (designated by us as "FHL5"). Lymphoblasts isolated from these patients had strongly decreased STXBP2 protein expression, and NK cells exhibited impaired cytotoxic granule exocytosis, a defect that could be overcome by ectopic expression of wild-type STXBP2. Furthermore, we provide evidence that syntaxin-11 is the main partner of STXBP2 in lymphocytes, as its expression required the presence of STXBP2. Our work shows that STXBP2 deficiency causes FHL5. These data indicate that STXBP2 is required at a late step of the secretory pathway for the release of cytotoxic granules by binding syntaxin 11, another component of the intracellular membrane fusion machinery.
Figures
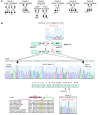
Figure 1. STXBP2 mutations in FHL5.
(A) Pedigrees from the FHL families with STXBP2 mutations. F3P1 and F3P2 belonged to the same tribe in Saudi Arabia. Consanguinity (double horizontal bars), affected individuals (black boxes and circles), carriers (half-filled boxes and circles), deceased individuals (diagonal bars) and subjects not available to participate in the study (asterisks) are indicated. (B) Positions of the 2 STXBP2 mutations. The splice mutation in intron 14 introduced 56 bp from the intronic sequence into the end of exon 15. The proline transition (P477L) occurred at a residue that had been conserved over evolution and within the members of the Munc18 family.
Figure 2. STXBP2 and STX11 expression in FHL5.
(A) Lymphoblasts from affected individuals showed normal levels of STXPB2 transcript. (B) STXBP2 transcript levels were quantified as the fold difference of mRNA levels for STXBP2 normalized to 18S, a housekeeping gene, in the lymphoblasts from control subjects and patients. Analyses were performed in triplicate. Data are mean ± SD. (C) Left: Western blot showing the expression of STXBP2, STX3, and STX11 proteins in lymphocytes from a healthy control, 3 STXBP2-deficient patients (F2P1, F6P1, and F5P1), and 2 STX11-deficient patients with nonsense mutations in STX11 located in the 5ι (P2) or 3ι (P1) part of the gene. Right: The abundance of STX3 and STX11 (relative to PI3K) via signal intensity measurement. Results are presented as the syntaxin/PI3K ratio, normalized against that obtained with a control (in arbitrary units).
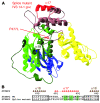
Figure 3. Model structure of STXBP2.
(A) A ribbon representation of the STXBP2 protein. By homology with STXBP1, the 3 domains are colored as follows: domain 1 in blue, domain 2 in green (with light and dark green indicating the 2 non-contiguous segments of the polypeptide chain that form this domain), domain 3a in yellow, and domain 3b in brown. The central cavity (formed by domains 1 and 3a) provides the binding surface for syntaxin. The mutated proline (P477L) and the α helix affected by the splice are highlighted in red. The IVS14-1G>C mutant induces aa replacements in the hydrophobic core. (B) Splice mutation, which replaces 17 aa of the WT sequence with 19 new residues. A remarkable conservation of a hydrophobic aa pattern can be observed. Hydrophobic aa are highlighted in green; the alternation of hydrophobic and hydrophilic aa every 3–4 residues is typical for amphipathic α helices.
Figure 4. Defective cytotoxic granule exocytosis of STXBP2-deficient NK lymphocytes.
(A) Exocytosis of cytotoxic granules from resting or STXBP2-deficient CD3–CD56+ NK cells cultured with IL-2 for 72 hours (F5P1, triangle and F5P2, diamond) compared with cells of healthy adult and infant donors (controls). Cytotoxic granule exocytosis (ΔCD107) represents the percentage of CD107+ NK cells stimulated with anti-CD16 and P815 cells subtracted from the percentage of CD107+ NK cells incubated with P815 cells alone. Horizontal bars represent mean values. **P <_ 0.01, control values compared with the median value for each patient. (**B**) CD107 expression of NK cells cultured with IL-2 for 15–20 days from either controls or STXBP2-deficient (F5P1, F5P2, and F6P1) individuals. Values represent mean (± SD) percentages of the CD107+ NK cells. (**C**) Cytotoxic granule exocytosis by T lymphoblasts. Induced CD107 surface expression on CD3+CD8+ T lymphoblasts from controls, STXBP2-deficient individuals (F2P1, F5P1, and F6P1), or STX11-deficient individuals (P1 and P2). Horizontal bars represent mean values. #_P > 0.1. (D) Restoration of the cytotoxic granule exocytosis in patient cells transfected with WT STXBP2 construct. PBMCs described in B were cotransfected with the WT STXBP2-FLAG–tagged vector or empty-FLAG–tagged vector and the insertless ECFP vector. For each individual, the ECFP-positive (transfected) and ECFP-negative (nontransfected) NK cell populations were tested as described in B. Dot plots were gated on CD3–CD56+ NK cells, and gates were set individually on the basis of NK cells incubated with P815 alone. Numbers indicate the percentage of degranulating NK cells. The results shown are representative of 2 independent experiments with similar results.
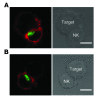
Figure 5. Normal polarization of cytotoxic granules in STXBP2-deficient NK cells.
Confocal microscopy of WT (A) and STXBP2-deficient (B) LAK cells conjugated with anti-CD16 P815 target cells. Cells were stained with perforin mAb (green) and phalloidin (red). Perforin polarized in 66% of control NK cell conjugates (n = 30) and 70% of STXBP2-deficient NK cells conjugates (n = 30). Scale bars: 5 μm. Data are representative of 3 independent experiments.
Similar articles
- Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion.
Spessott WA, Sanmillan ML, McCormick ME, Patel N, Villanueva J, Zhang K, Nichols KE, Giraudo CG. Spessott WA, et al. Blood. 2015 Mar 5;125(10):1566-77. doi: 10.1182/blood-2014-11-610816. Epub 2015 Jan 6. Blood. 2015. PMID: 25564401 Free PMC article. - Familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) is caused by mutations in Munc18-2 and impaired binding to syntaxin 11.
zur Stadt U, Rohr J, Seifert W, Koch F, Grieve S, Pagel J, Strauss J, Kasper B, Nürnberg G, Becker C, Maul-Pavicic A, Beutel K, Janka G, Griffiths G, Ehl S, Hennies HC. zur Stadt U, et al. Am J Hum Genet. 2009 Oct;85(4):482-92. doi: 10.1016/j.ajhg.2009.09.005. Am J Hum Genet. 2009. PMID: 19804848 Free PMC article. - Novel STXBP2 mutation causing familial hemophagocytic lymphohistiocytosis.
Jain R, Puliyel M, Moses PD, Sieni E. Jain R, et al. Indian Pediatr. 2012 Jun;49(6):488-90. doi: 10.1007/s13312-012-0094-5. Indian Pediatr. 2012. PMID: 22796692 - Angeborene hämophagozytische Lymphohistiozytose (HLH).
Pachlopnik Schmid J, de Saint Basile G. Pachlopnik Schmid J, et al. Klin Padiatr. 2010 Nov;222(6):345-50. doi: 10.1055/s-0029-1246165. Epub 2010 May 10. Klin Padiatr. 2010. PMID: 20458667 Review. - A unique SNARE machinery for exocytosis of cytotoxic granules and platelets granules.
Tang BL. Tang BL. Mol Membr Biol. 2015;32(4):120-6. doi: 10.3109/09687688.2015.1079934. Mol Membr Biol. 2015. PMID: 26508555 Review.
Cited by
- Munc18b/STXBP2 is required for platelet secretion.
Al Hawas R, Ren Q, Ye S, Karim ZA, Filipovich AH, Whiteheart SW. Al Hawas R, et al. Blood. 2012 Sep 20;120(12):2493-500. doi: 10.1182/blood-2012-05-430629. Epub 2012 Jul 12. Blood. 2012. PMID: 22791290 Free PMC article. - Splicing factor SRSF1 is essential for CD8 T cell function and host antigen-specific viral immunity.
Juarez I, Su S, Herbert ZT, Teijaro JR, Moulton VR. Juarez I, et al. Front Immunol. 2022 Sep 16;13:906355. doi: 10.3389/fimmu.2022.906355. eCollection 2022. Front Immunol. 2022. PMID: 36189299 Free PMC article. - Understanding inborn errors of immunity: A lens into the pathophysiology of monogenic inflammatory bowel disease.
Ouahed JD. Ouahed JD. Front Immunol. 2022 Sep 29;13:1026511. doi: 10.3389/fimmu.2022.1026511. eCollection 2022. Front Immunol. 2022. PMID: 36248828 Free PMC article. Review. - Molecular Classification of Primary Immunodeficiencies of T Lymphocytes.
Comrie WA, Lenardo MJ. Comrie WA, et al. Adv Immunol. 2018;138:99-193. doi: 10.1016/bs.ai.2018.02.003. Epub 2018 Mar 29. Adv Immunol. 2018. PMID: 29731008 Free PMC article. Review. - Two Brothers with Atypical _UNC13D-_Related Hemophagocytic Lymphohistiocytosis Characterized by Massive Lung and Brain Involvement.
Giardino G, De Luca M, Cirillo E, Palma P, Romano R, Valeriani M, Papetti L, Saunders C, Cancrini C, Pignata C. Giardino G, et al. Front Immunol. 2017 Dec 21;8:1892. doi: 10.3389/fimmu.2017.01892. eCollection 2017. Front Immunol. 2017. PMID: 29312353 Free PMC article.
References
- Henter J.I., Elinder G., Ost A. Diagnostic guidelines for hemophagocytic lymphohistiocytosis. The FHL Study Group of the Histiocyte Society. Semin. Oncol. 1991;18:29–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases