An oestrogen-receptor-alpha-bound human chromatin interactome - PubMed (original) (raw)
. 2009 Nov 5;462(7269):58-64.
doi: 10.1038/nature08497.
Mei Hui Liu, You Fu Pan, Jun Liu, Han Xu, Yusoff Bin Mohamed, Yuriy L Orlov, Stoyan Velkov, Andrea Ho, Poh Huay Mei, Elaine G Y Chew, Phillips Yao Hui Huang, Willem-Jan Welboren, Yuyuan Han, Hong Sain Ooi, Pramila N Ariyaratne, Vinsensius B Vega, Yanquan Luo, Peck Yean Tan, Pei Ye Choy, K D Senali Abayratna Wansa, Bing Zhao, Kar Sian Lim, Shi Chi Leow, Jit Sin Yow, Roy Joseph, Haixia Li, Kartiki V Desai, Jane S Thomsen, Yew Kok Lee, R Krishna Murthy Karuturi, Thoreau Herve, Guillaume Bourque, Hendrik G Stunnenberg, Xiaoan Ruan, Valere Cacheux-Rataboul, Wing-Kin Sung, Edison T Liu, Chia-Lin Wei, Edwin Cheung, Yijun Ruan
Affiliations
- PMID: 19890323
- PMCID: PMC2774924
- DOI: 10.1038/nature08497
An oestrogen-receptor-alpha-bound human chromatin interactome
Melissa J Fullwood et al. Nature. 2009.
Abstract
Genomes are organized into high-level three-dimensional structures, and DNA elements separated by long genomic distances can in principle interact functionally. Many transcription factors bind to regulatory DNA elements distant from gene promoters. Although distal binding sites have been shown to regulate transcription by long-range chromatin interactions at a few loci, chromatin interactions and their impact on transcription regulation have not been investigated in a genome-wide manner. Here we describe the development of a new strategy, chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) for the de novo detection of global chromatin interactions, with which we have comprehensively mapped the chromatin interaction network bound by oestrogen receptor alpha (ER-alpha) in the human genome. We found that most high-confidence remote ER-alpha-binding sites are anchored at gene promoters through long-range chromatin interactions, suggesting that ER-alpha functions by extensive chromatin looping to bring genes together for coordinated transcriptional regulation. We propose that chromatin interactions constitute a primary mechanism for regulating transcription in mammalian genomes.
Figures
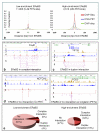
Figure 1. ChIA-PET method with validations
(a) ChIA-PET schematic. DNA fragments in sonicated, ChIP-enriched chromatin complexes were processed by linker ligation, proximity ligation, PET extraction, sequencing, and mapping to reveal interacting loci. (b) ChIA-PET browser tracks: 1, H3K4me3 ChIP-Seq; 2, RNAPII ChIP-Seq; 3, ERα (orange) and FoxA1 ChIP-chip (green); 4, ERα ChIA-PET density ; 5, Inter-ligation PETs. Inset: 3C validation of interacting ERαBS (purple) and controls (blue) under ethanol (ET) and oestrogen-induction (E2). (c) 4C validation, showing 4C bait region (blue) and interaction targets (purple bars). (d) FISH validation, showing increased P2/P1 interactions under E2-induction with background normalization (P3/P2). FISH probe genomic locations (P1/P2/P3) are indicated.
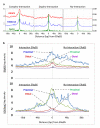
Figure 2. ERαBS reproducibility and association with chromatin interactions
(a) Numbers of ERαBS identified with different ChIP enrichment cutoffs and reproducibility analyses as measured by overlapping with another ChIA-PET dataset (IHH015F), ChIP-Seq, and ChIP-chip data. Examples of ERαBS involved in (b) complex-interactions, (c) duplex-interactions, and (d) no-interactions (singleton inter-ligation PETs only or no inter-ligation PETs). (e) ERαBS distribution in different categories of interactions as exemplified in b-d.
Figure 3. Association of ERα-bound chromatin interactions with functional marks
(a) Association of ERαBS in complex-, duplex-, and no-interaction categories with RNAPII (red), H3K4me3 (blue), and FoxA1 (green) functional marks. (b) Association of proximal and distal interacting and non-interacting ERαBS with H3K4me3 and RNAPII functional marks.
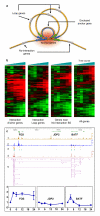
Figure 4. Proposed ERα-bound chromatin interaction and transcription regulation mechanism
(a) Distal ERαBS interact with proximal sites, forming chromatin loops. Anchor genes (green and blue) are close to interaction anchors with concentrated active transcriptional machinery (red shading). Other genes far from interaction centers (grey) are less active. (b) Expression microarray data (oestrogen induction from 0 to 48h; red denotes activation; green denotes repression) for interaction anchor genes, loop genes, and genes near no-interaction ERαBS, with all other UCSC Genes (“All genes”) denoting background. (c) ChIA-PET interactions data at the FOS/JDP2/BATF loci. Transcription activities are shown by H3K4me3/RNAPII ChIP-Seq and RT-qPCR analysis (oestrogen induction from 0 to 24h).
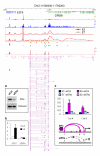
Figure 5. ERα-bound chromatin interactions are required for transcription activation
Genome browser at the GREB1 locus showing data tracks: H3K4me3 and RNAPII ChIP-Seq (1 and 2); RNAPII ChIP-qPCR scans (3 and 4) using different RNAPII antibodies under oestrogen-induction (E2, in red) and ethanol control (ET, in grey); ERα (orange) and FoxA1 (green) ChIP-chip (5); ChIA-PET density (6) and interaction data (7). Inset: siRNA knockdown experiments. MCF-7 cells were transfected with siRNA against ERα (siERα) or control (siCtrl), and then analyzed by (a) western blot with ERα and calnexin (control) antibodies; (b) RT-qPCR on GREB1 expression; and (c) 3C assays at GREB1: siERα knockdown abolishes chromatin interactions and turns off transcription.
Similar articles
- AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription.
Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E. Tan SK, et al. EMBO J. 2011 May 13;30(13):2569-81. doi: 10.1038/emboj.2011.151. EMBO J. 2011. PMID: 21572391 Free PMC article. - Chromatin Interaction Analysis with Paired-End Tag Sequencing (ChIA-PET) for mapping chromatin interactions and understanding transcription regulation.
Goh Y, Fullwood MJ, Poh HM, Peh SQ, Ong CT, Zhang J, Ruan X, Ruan Y. Goh Y, et al. J Vis Exp. 2012 Apr 30;(62):3770. doi: 10.3791/3770. J Vis Exp. 2012. PMID: 22564980 Free PMC article. - Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters.
Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Kininis M, et al. Mol Cell Biol. 2007 Jul;27(14):5090-104. doi: 10.1128/MCB.00083-07. Epub 2007 May 21. Mol Cell Biol. 2007. PMID: 17515612 Free PMC article. - Estrogen receptor-mediated long-range chromatin interactions and transcription in breast cancer.
Liu MH, Cheung E. Liu MH, et al. Mol Cell Endocrinol. 2014 Jan 25;382(1):624-632. doi: 10.1016/j.mce.2013.09.019. Epub 2013 Sep 24. Mol Cell Endocrinol. 2014. PMID: 24071518 Review. - SATB1-mediated functional packaging of chromatin into loops.
Kohwi-Shigematsu T, Kohwi Y, Takahashi K, Richards HW, Ayers SD, Han HJ, Cai S. Kohwi-Shigematsu T, et al. Methods. 2012 Nov;58(3):243-54. doi: 10.1016/j.ymeth.2012.06.019. Epub 2012 Jul 7. Methods. 2012. PMID: 22782115 Free PMC article. Review.
Cited by
- Challenges and Opportunities in Understanding Genetics of Fungal Diseases: Towards a Functional Genomics Approach.
Bruno M, Matzaraki V, van de Veerdonk FL, Kumar V, Netea MG. Bruno M, et al. Infect Immun. 2021 Jul 15;89(8):e0000521. doi: 10.1128/IAI.00005-21. Epub 2021 Jul 15. Infect Immun. 2021. PMID: 34031131 Free PMC article. Review. - Comparison of the Hi-C, GAM and SPRITE methods using polymer models of chromatin.
Fiorillo L, Musella F, Conte M, Kempfer R, Chiariello AM, Bianco S, Kukalev A, Irastorza-Azcarate I, Esposito A, Abraham A, Prisco A, Pombo A, Nicodemi M. Fiorillo L, et al. Nat Methods. 2021 May;18(5):482-490. doi: 10.1038/s41592-021-01135-1. Epub 2021 May 7. Nat Methods. 2021. PMID: 33963348 Free PMC article. - PredTAD: A machine learning framework that models 3D chromatin organization alterations leading to oncogene dysregulation in breast cancer cell lines.
Chyr J, Zhang Z, Chen X, Zhou X. Chyr J, et al. Comput Struct Biotechnol J. 2021 May 7;19:2870-2880. doi: 10.1016/j.csbj.2021.05.013. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34093998 Free PMC article. - The second decade of 3C technologies: detailed insights into nuclear organization.
Denker A, de Laat W. Denker A, et al. Genes Dev. 2016 Jun 15;30(12):1357-82. doi: 10.1101/gad.281964.116. Genes Dev. 2016. PMID: 27340173 Free PMC article. Review. - Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression.
Xu Y, Guo W, Li P, Zhang Y, Zhao M, Fan Z, Zhao Z, Yan J. Xu Y, et al. PLoS Genet. 2016 May 2;12(5):e1005992. doi: 10.1371/journal.pgen.1005992. eCollection 2016 May. PLoS Genet. 2016. PMID: 27135601 Free PMC article.
References
- Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–5. - PubMed
- Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci. 2008;13:929–43. - PubMed
- Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. - PubMed
- Wold B, Myers RM. Sequence census methods for functional genomics. Nat Methods. 2008;5:19–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 HG004557/HG/NHGRI NIH HHS/United States
- R01HG004456-01/HG/NHGRI NIH HHS/United States
- 1U54HG004557-01/HG/NHGRI NIH HHS/United States
- R01 HG004456/HG/NHGRI NIH HHS/United States
- R01 HG003521/HG/NHGRI NIH HHS/United States
- R01HG003521-01/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases