Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis - PubMed (original) (raw)
. 2009 Dec;41(12):1295-302.
doi: 10.1038/ng.476. Epub 2009 Nov 8.
Cristina Cebrian, Xuan Chi, Satu Kuure, Richard Kuo, Carlton M Bates, Silvia Arber, John Hassell, Lesley MacNeil, Masato Hoshi, Sanjay Jain, Naoya Asai, Masahide Takahashi, Kai M Schmidt-Ott, Jonathan Barasch, Vivette D'Agati, Frank Costantini
Affiliations
- PMID: 19898483
- PMCID: PMC2787691
- DOI: 10.1038/ng.476
Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis
Benson C Lu et al. Nat Genet. 2009 Dec.
Erratum in
- Nat Genet. 2010 Apr;42(4):361
Abstract
Glial cell line-derived neurotrophic factor signaling through the Ret receptor tyrosine kinase is crucial for ureteric bud branching morphogenesis during kidney development, yet few of the downstream genes are known. Here we show that the ETS transcription factors Etv4 and Etv5 are positively regulated by Ret signaling in the ureteric bud tips. Mice lacking both Etv4 alleles and one Etv5 allele show either renal agenesis or severe hypodysplasia, whereas kidney development fails completely in double homozygotes. We identified several genes whose expression in the ureteric bud depends on Etv4 and Etv5, including Cxcr4, Myb, Met and Mmp14. Thus, Etv4 and Etv5 are key components of a gene network downstream of Ret that promotes and controls renal branching morphogenesis.
Conflict of interest statement
Competing Financial Interests
The authors have no competing financial interests.
Figures
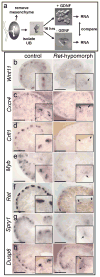
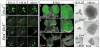
Figure 1. A screen for genes upregulated by GDNF/Ret signaling in the ureteric bud
a, E11.5 UBs isolated from mesenchyme were cultured with or without GDNF. RNA was analyzed with Affymetrix U74Av2 and 430A arrays (Table 1 and Supplementary Tables 1 and 2). b–h, in situ hybridization to control or Rettm2(RET)Vpa/Rettm2(RET)Vpa kidneys (E14.5–E15.5). Unlike _Ret_−/− mice, which usually have renal agenesis, Rettm2(RET)Vpa homozygotes (expressing only the Ret51 isoform) have a milder phenotype with reduced branching. Thus, the Rettm2(RET)Vpa allele behaves as a hypomorph due to decreased Ret signaling activity. Scale bars 100 μM. Insets, 2x enlargements of UB tips (arrows).
Figure 2. Similar expression patterns of Etv4 and Etv5 during kidney development
a–f, expression of Etv4-lacZ and Etv5-lacZ at E9.5–11.5 in WD (white arrows) and UB (black arrows). g–h, Etv4 and Etv5 ISH at E11.5. Arrows, UB; dotted lines delineate MM. i–j, Etv4 and Etv5 ISH in E14.5 kidneys. Arrows, UB tips; arrowheads, MM; open arrows, nascent nephrons. k, Etv4 protein expression (red) in UB tips (arrows), MM (arrowheads) and nascent nephrons (open arrows); l, lack of Etv4 staining in _Etv4_−/− kidney shows antibody specificity. Scale bars 100 μM except for k,l (50 μM).
Figure 3. GDNF/Ret signaling regulates Etv4 and Etv5 expression in UB tips
a–b, Increased expression of Etv4-lacZ and Etv5-lacZ in the vicinity of GDNF-soaked beads (black asterisks) but not control BSA-soaked beads (red asterisk). E15.5 (a) or E13.5 kidneys (b) were cultured for 48 hrs. c–f, Greatly reduced expression of Etv4 and Etv5 mRNAs in UB tips (arrows) of _Ret_-hypomorphic E14.5 kidneys (e, f) compared to WT kidneys (c, d). However, expression in nascent nephrons (arrowheads) persists in the mutant, consistent with the lack of Ret expression in the developing nephrons. Insets in c–f, enlargement of UB tips. Scale bars 100 μM.
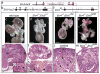
Figure 4. Severe renal developmental defects in compound Etv4/Etv5 mutant newborn mice with two different Etv5 knockout alleles
a–b, Etv4-lacZ and Etv5-lacZ alleles. c–h, renal defects in Etv4_−/−;Etv5_+/− compound mutants. c, normal kidneys (k) in WT. d, unilateral renal agenesis and unilateral hypodysplasia in Etv4_−/−;Etv5_+/− mutant. e–f, histology of WT and Etv4_−/−;Etv5_+/− kidneys. m, medulla; c, cortex; n, nephrogenic zone. g–h, enlargements of glomeruli. Note the small size, absent nephrogenic zone, sparse but apparently normal glomeruli, and multiple cysts in mutant kidney. i, Etv5M allele. Exons 2–5, including initiation codon, are deleted. j–m, renal defects in compound mutants with Etv5M allele. j, Etv4_−/−;Etv5M/+_ mutant with bilateral hypoplasia. k, Etv4_−/−;Etv5M/M_ double homozygote with one ureter and tiny kidney rudiment. l, histology of normal kidney, with numerous glomeruli (arrows). m, histology of the Etv4_−/−;Etv5M/M_ kidney rudiment in k, with few epithelial elements and no glomeruli. Scale bars:1 mm in c, d, j, k; 25 μM in other panels.
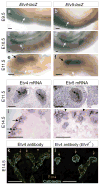
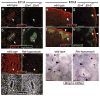
Figure 5. Ureteric bud branching defects in Etv4_−/−;Etv5_+/− compoundmutants
a–d, culture of one control (a) and three representative compound mutant E11.5 kidney primordia, all carrying Hoxb7/myrVenus. Mutant b has a WD (arrowhead) but fails to make a UB, c has a UB (arrow) that fails to branch, d has a normal T-shaped UB at E11.5 but branches less than control when cultured. e–j, Hoxb7/myrVenus fluorescence reveals normal UB branching in a control kidney at E16.5 (e–f), reduced and irregular branching in the hypoplastic kidneys of one mutant (g–h), and a single unbranched ureter with no kidney in another mutant (i–j). Boxed areas are enlarged in f, h, j. k–m, culture of isolated, mesenchyme-free UBs in Matrigel. The Etv4+/− (k) and Etv4_−/− (l) UBs grow and branch extensively but the Etv4_−/−;Etv5+/− (m) does so only slightly. Scale bars 100 μM (a–d, k–m), 500 μM (e–j).
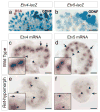
Figure 6. Gene expression in UB tips of Etv4_−/−;Etv5_+/− compoundmutant kidneys
a–d, Expression of Ret, Crlf, Dusp6 and Etv5 mRNAs was similar in the UB tips of mutant and control kidneys at E15.5, although Ret was reduced in some tips. e–h, At E15.5, expression of Wnt11, Cxcr4, Myb and Met was greatly reduced in mutant kidneys. i–l, At E13.5, Wnt11 expression was normal, but Cxcr4, Myb and Met were greatly reduced. Insets show 2x enlargements of UB tips. Black arrows indicate tips with normal expression, white arrows those with reduced expression, and asterisks indicate nascent nephrons. Controls include WT, Etv4+/− or Etv5+/−. Scale bars 100 μM.
Figure 7. Reduced MMP14 expression in Etv4_−/−;Etv5_+/− compoundmutant and _Ret_-hypomorphic kidneys
a–h, MMP14 protein in WT and Etv4_−/−;Etv5_+/− kidneys. At both E11.5 and E15.5, MMP14 is detected in UB epithelium (arrows) and surrounding mesenchyme of WT kidneys (a–b, e–f) but is greatly reduced in UB and MM of Etv4_−/−;Etv5_+/− kidneys (c–d, g–h). The UB is marked by Hoxb7/myrVenus in b, d, f, h. In the Etv4_−/−;Etv5_+/− at E15.5 (g–h), strong MMP14 expression persists in the peripheral stroma (arrowheads). i–n, analysis of _Ret_-hypomorphic kidneys. MMP14 protein (i,k) is reduced in the UB epithelium and the surrounding MM of _Ret_-hypomorphic kidneys at E11.5. In WT E15.5 kidney (m), Mmp14 mRNA is detected in UB epithelium (arrows) and stroma (arrowheads) but not in cap mesenchyme surrounding UB tips (asterisks). In _Ret_-hypomorphic kidney (n), Mmp14 mRNA is reduced in UB epithelium but persists in stroma (cap mesenchyme is reduced in quantity in the mutant). Scale bars 50 μM.
Similar articles
- GDNF/Ret signaling and renal branching morphogenesis: From mesenchymal signals to epithelial cell behaviors.
Costantini F. Costantini F. Organogenesis. 2010 Oct-Dec;6(4):252-62. doi: 10.4161/org.6.4.12680. Organogenesis. 2010. PMID: 21220964 Free PMC article. - The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development.
Kuure S, Chi X, Lu B, Costantini F. Kuure S, et al. Development. 2010 Jun;137(12):1975-9. doi: 10.1242/dev.051656. Epub 2010 May 12. Development. 2010. PMID: 20463033 Free PMC article. - Kidney development in the absence of Gdnf and Spry1 requires Fgf10.
Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, Costantini F. Michos O, et al. PLoS Genet. 2010 Jan 15;6(1):e1000809. doi: 10.1371/journal.pgen.1000809. PLoS Genet. 2010. PMID: 20084103 Free PMC article. - GDNF/Ret signaling and the development of the kidney.
Costantini F, Shakya R. Costantini F, et al. Bioessays. 2006 Feb;28(2):117-27. doi: 10.1002/bies.20357. Bioessays. 2006. PMID: 16435290 Review. - Renin-angiotensin system-growth factor cross-talk: a novel mechanism for ureteric bud morphogenesis.
Yosypiv IV. Yosypiv IV. Pediatr Nephrol. 2009 Jun;24(6):1113-20. doi: 10.1007/s00467-008-1021-9. Epub 2008 Oct 29. Pediatr Nephrol. 2009. PMID: 18958502 Free PMC article. Review.
Cited by
- Multifunctional extracellular vesicles and edaravone-loaded scaffolds for kidney tissue regeneration by activating GDNF/RET pathway.
Lee SY, Park JM, Rhim WK, Lee EH, Lee SH, Kim JY, Cha SG, Lee SH, Kim B, Hwang DY, Rho S, Ahn TK, Kim BS, Han DK. Lee SY, et al. Nano Converg. 2024 Oct 26;11(1):43. doi: 10.1186/s40580-024-00450-5. Nano Converg. 2024. PMID: 39460807 Free PMC article. - GDNF promotes hair formation and cutaneous wound healing by targeting bulge stem cells.
Lisse TS, Sharma M, Vishlaghi N, Pullagura SR, Braun RE. Lisse TS, et al. NPJ Regen Med. 2020 Jun 12;5:13. doi: 10.1038/s41536-020-0098-z. eCollection 2020. NPJ Regen Med. 2020. PMID: 32566252 Free PMC article. - Deletion of the prorenin receptor from the ureteric bud causes renal hypodysplasia.
Song R, Preston G, Ichihara A, Yosypiv IV. Song R, et al. PLoS One. 2013 May 21;8(5):e63835. doi: 10.1371/journal.pone.0063835. Print 2013. PLoS One. 2013. PMID: 23704941 Free PMC article. - Non-muscle myosin II deletion in the developing kidney causes ureter-bladder misconnection and apical extrusion of the nephric duct lineage epithelia.
Haque F, Kaku Y, Fujimura S, Ohmori T, Adelstein RS, Nishinakamura R. Haque F, et al. Dev Biol. 2017 Jul 1;427(1):121-130. doi: 10.1016/j.ydbio.2017.04.020. Epub 2017 May 3. Dev Biol. 2017. PMID: 28478097 Free PMC article. - Postnatal prolongation of mammalian nephrogenesis by excess fetal GDNF.
Li H, Kurtzeborn K, Kupari J, Gui Y, Siefker E, Lu B, Mätlik K, Olfat S, Montaño-Rodríguez AR, Huh SH, Costantini F, Andressoo JO, Kuure S. Li H, et al. Development. 2021 May 15;148(10):dev197475. doi: 10.1242/dev.197475. Epub 2021 May 25. Development. 2021. PMID: 34032268 Free PMC article.
References
- Costantini F. Renal branching morphogenesis: concepts, questions, and recent advances. Differentiation. 2006;74:402–21. - PubMed
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–29. - PubMed
- Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet. 2007;8:791–802. - PubMed
- Saxen L. Organogenesis of the Kidney. Cambridge University Press; Cambridge: 1987.
- Hoy WE, et al. Nephron number, glomerular volume, renal disease and hypertension. Curr Opin Nephrol Hypertens. 2008;17:258–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK55388/DK/NIDDK NIH HHS/United States
- R01 DK075578/DK/NIDDK NIH HHS/United States
- R01 DK082531/DK/NIDDK NIH HHS/United States
- P01 DK055388/DK/NIDDK NIH HHS/United States
- DK075578/DK/NIDDK NIH HHS/United States
- P01 DK055388-08/DK/NIDDK NIH HHS/United States
- R01 DK082531-01/DK/NIDDK NIH HHS/United States
- T35 DK093430/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous