Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor - PubMed (original) (raw)
Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor
Kailang Wu et al. Proc Natl Acad Sci U S A. 2009.
Abstract
NL63 coronavirus (NL63-CoV), a prevalent human respiratory virus, is the only group I coronavirus known to use angiotensin-converting enzyme 2 (ACE2) as its receptor. Incidentally, ACE2 is also used by group II SARS coronavirus (SARS-CoV). We investigated how different groups of coronaviruses recognize the same receptor, whereas homologous group I coronaviruses recognize different receptors. We determined the crystal structure of NL63-CoV spike protein receptor-binding domain (RBD) complexed with human ACE2. NL63-CoV RBD has a novel beta-sandwich core structure consisting of 2 layers of beta-sheets, presenting 3 discontinuous receptor-binding motifs (RBMs) to bind ACE2. NL63-CoV and SARS-CoV have no structural homology in RBD cores or RBMs; yet the 2 viruses recognize common ACE2 regions, largely because of a "virus-binding hotspot" on ACE2. Among group I coronaviruses, RBD cores are conserved but RBMs are variable, explaining how these viruses recognize different receptors. These results provide a structural basis for understanding viral evolution and virus-receptor interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
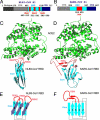
Fig. 1.
Structure of NL63-CoV RBD complexed with human ACE2. (A) Domain structure of the NL63-CoV spike protein. Unique, unique region; NTR, N-terminal region; Central, central region; CTR, C-terminal region; HR-N, heptad-repeat N; HR-C, heptad-repeat C; TM, transmembrane anchor; IC, intracellular tail. The unique domain only exists in NL63-CoV and is not involved in receptor binding (19, 20). (B) Overall structure of NL63-CoV RBD complexed with human ACE2. The RBD core is in cyan, RBMs in red, and ACE2 in green. (C) Averaged electron density map contoured at 1.0 σ and covering a portion of the NL63-CoV–ACE2 interface. (D) Sequence and secondary structures of NL63-CoV RBD. Beta-strands are drawn as arrows. RBMs are in red; the remainder of the RBD is in cyan. Disordered regions are shown as dashed lines. (E) Kinetics and binding affinity of NL63-CoV RBD and human ACE2 by surface plasmon resonance using Biacore. Structural illustrations were made using Povscript (31).
Fig. 2.
Structural comparison of NL63-CoV and SARS-CoV RBDs. (A) Domain structure of NL63-CoV S1. The boundaries of the RBD were determined by limited proteolysis of longer S1 fragments, followed by N-terminal sequencing and mass spectrometric analysis of the digestion fragments. The RBMs were identified from the crystal structure of the NL63-CoV RBD in complex with ACE2. (B) Domain structure of SARS-CoV S1 (5). (C) Another view of the structure of the NL63-CoV-RBD–ACE2 complex, which is derived by rotating the one in Fig. 1_B_ by 90° clockwise along a vertical axis. (D) Structure of the SARS-CoV-RBD–ACE2 complex (PDB 2AJF) (5), from the same orientation as in C. (E) Schematic illustration of the topology of NL63-CoV RBD. Strands are drawn as arrows. (F) Schematic illustration of the topology of SARS-CoV RBD.
Fig. 3.
Structural features of NL63-CoV RBD and RBMs. (A) Beta-sandwich core structure of NL63-CoV RBD. Hydrophobic residues between β-sheet layers are in yellow, cysteines in blue, glycans in green, and glycosylated asparagines in magenta. There exist 3 predicted N-linked glycosylation sites in the RBD (Asn-486, Asn-506, and Asn-512), and 2 of them are confirmed in the structure (Asn-486 and Asn-512). (B) NL63-CoV RBMs. (C) Residues on NL63-CoV RBMs that directly contact ACE2.
Fig. 4.
Structural comparison of NL63-CoV–ACE2 and SARS-CoV–ACE2 interfaces. (A) Enlarged view of the NL63-CoV–ACE2 interface, from the same orientation as in Fig. 2_C_. VBMs on ACE2 are in blue, and RBMs on NL63-CoV are in red. Arrow indicates the bowl-shaped cavity surrounded by 3 viral RBMs. (B) Enlarged view of the SARS-CoV–ACE2 interface, from the same orientation as in A. (C) Footprint of NL63-CoV on the surface of ACE2. The view is derived from the one in A by rotating ACE2 by 90° along a horizontal axis, in such a way that the edge facing the viewer moves up. VBM1 residues are in orange, VBM2 residues in magenta, and VBM3 residues in red. (D) Footprint of SARS-CoV on the surface of ACE2, from the same orientation as in C. VBM1b residues are in green.
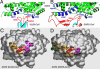
Fig. 5.
A common virus-binding hotspot on ACE2 for the binding of both NL63-CoV and SARS-CoV. (A) Stick-and-ball representation of the hotspot at the NL63-CoV–ACE2 interface. (B) Corey-Pauling-Koltun representation of the hotspot at the NL63-CoV–ACE2 interface. (C) Stick-and-ball representation of the hotspot at the SARS-CoV–ACE2 interface. (D) Corey-Pauling-Koltun representation of the hotspot at the SARS-CoV–ACE2 interface.
Similar articles
- Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction.
Lin HX, Feng Y, Wong G, Wang L, Li B, Zhao X, Li Y, Smaill F, Zhang C. Lin HX, et al. J Gen Virol. 2008 Apr;89(Pt 4):1015-1024. doi: 10.1099/vir.0.83331-0. J Gen Virol. 2008. PMID: 18343844 - Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections.
Li F. Li F. J Virol. 2008 Jul;82(14):6984-91. doi: 10.1128/JVI.00442-08. Epub 2008 Apr 30. J Virol. 2008. PMID: 18448527 Free PMC article. - Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors.
Hofmann H, Simmons G, Rennekamp AJ, Chaipan C, Gramberg T, Heck E, Geier M, Wegele A, Marzi A, Bates P, Pöhlmann S. Hofmann H, et al. J Virol. 2006 Sep;80(17):8639-52. doi: 10.1128/JVI.00560-06. J Virol. 2006. PMID: 16912312 Free PMC article. - Insights from the association of SARS-CoV S-protein with its receptor, ACE2.
Li W, Choe H, Farzan M. Li W, et al. Adv Exp Med Biol. 2006;581:209-18. doi: 10.1007/978-0-387-33012-9_36. Adv Exp Med Biol. 2006. PMID: 17037532 Free PMC article. Review. No abstract available. - Attachment factor and receptor engagement of SARS coronavirus and human coronavirus NL63.
Hofmann H, Marzi A, Gramberg T, Geier M, Pyrc K, van der Hoek L, Berkhout B, Pöhlmann S. Hofmann H, et al. Adv Exp Med Biol. 2006;581:219-27. doi: 10.1007/978-0-387-33012-9_37. Adv Exp Med Biol. 2006. PMID: 17037533 Free PMC article. Review. No abstract available.
Cited by
- Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo-electron microscopy.
Walls A, Tortorici MA, Bosch BJ, Frenz B, Rottier PJ, DiMaio F, Rey FA, Veesler D. Walls A, et al. Protein Sci. 2017 Jan;26(1):113-121. doi: 10.1002/pro.3048. Epub 2016 Oct 18. Protein Sci. 2017. PMID: 27667334 Free PMC article. - The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity.
Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB. Ovsyannikova IG, et al. Immunol Rev. 2020 Jul;296(1):205-219. doi: 10.1111/imr.12897. Epub 2020 Jul 13. Immunol Rev. 2020. PMID: 32658335 Free PMC article. Review. - Structural Analysis of the OC43 Coronavirus 2'-O-RNA Methyltransferase.
Dostalik P, Krafcikova P, Silhan J, Kozic J, Chalupska D, Chalupsky K, Boura E. Dostalik P, et al. J Virol. 2021 Jul 12;95(15):e0046321. doi: 10.1128/JVI.00463-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011548 Free PMC article. - Identification of SARS-CoV-2 Cell Entry Inhibitors by Drug Repurposing Using in silico Structure-Based Virtual Screening Approach.
Choudhary S, Malik YS, Tomar S. Choudhary S, et al. Front Immunol. 2020 Jul 10;11:1664. doi: 10.3389/fimmu.2020.01664. eCollection 2020. Front Immunol. 2020. PMID: 32754161 Free PMC article. - Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
Niu S, Zhao Z, Liu Z, Rong X, Chai Y, Bai B, Han P, Shang G, Ren J, Wang Y, Zhao X, Liu K, Tian W-x, Wang Q, Gao GF. Niu S, et al. J Virol. 2024 Mar 19;98(3):e0115723. doi: 10.1128/jvi.01157-23. Epub 2024 Feb 2. J Virol. 2024. PMID: 38305152 Free PMC article.
References
- Baranowski E, Ruiz-Jarabo CM, Domingo E. Evolution of cell recognition by viruses. Science. 2001;292:1102–1105. - PubMed
- Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. - PubMed
- Carfi A, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. - PubMed
- Li F, Li WH, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous