A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development - PubMed (original) (raw)
A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development
Xin-Jian He et al. Genes Dev. 2009.
Abstract
RNA-directed DNA methylation (RdDM) is a conserved mechanism for epigenetic silencing of transposons and other repetitive elements. We report that the rdm4 (RNA-directed DNA Methylation4) mutation not only impairs RdDM, but also causes pleiotropic developmental defects in Arabidopsis. Both RNA polymerase II (Pol II)- and Pol V-dependent transcripts are affected in the rdm4 mutant. RDM4 encodes a novel protein that is conserved from yeast to humans and interacts with Pol II and Pol V in plants. Our results suggest that RDM4 functions in epigenetic regulation and plant development by serving as a transcriptional regulator for RNA Pol V and Pol II, respectively.
Figures
Figure 1.
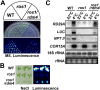
TGS phenotypes of the ros1rdm4 mutant plants. (A,B) Expression of the RD29A-LUC transgene by luminescence emission. (A) Wild type, ros1, and ros1rdm4 were grown on MS plates and imaged after cold treatment (24 h, 4°C). (B) The indicated plants were treated with 200 mM NaCl for 3 h, followed by luminescence imaging. (C) The transcript levels of endogenous RD29A, and RD29A-LUC and 35S-NPTII transgenes in wild type, ros1, and ros1rdm4 were determined by RNA blot analysis. 18S rRNA hybridization and ethidium bromide-stained rRNA bands were used as RNA loading controls, and COR15A was used as a cold treatment control.
Figure 2.
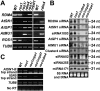
Effect of rdm4 on DNA methylation. The percentage of cytosine methylation at transgene (A) and endogenous (B) RD29A promoters was determined by bisulfite sequencing. The percentage of cytosine methylation on CG, CHG, and CHH (H stands for A, T, or C) sites is shown. (C) The rdm4 mutation reduces DNA methylation at the endogenous RD29A promoter as determined by Southern hybridization. Genomic DNA was digested with the methylation-sensitive restriction enzyme BstUI. (D) The rdm4 mutation reduces DNA methylation at 5S rDNA repeat. (E) Genomic DNA from indicated genotypes was digested with the methylation-sensitive restriction enzyme HaeIII, followed by Southern hybridization with the AtMU1 probe. (F) Effect of rdm4 on DNA methylation of AtSN1. PCR amplification of AtSN1 was performed after the genomic DNA was digested with HaeIII. TUB8 DNA was amplified as an internal control.
Figure 3.
Effect of the rdm4 mutation on RNA transcript levels and siRNA accumulation. (A) The RNA transcript levels of the indicated loci were determined by semiquantitative RT–PCR. TUB8 was used as an internal control. (B) Detection of small RNAs in the indicated genotypes by Northern blot analysis. The ethidium bromide-stained gel corresponding to 5S rRNA and tRNA was included as a loading control. The positions of size markers are indicated (24 nt or 21 nt). (C) RT–PCR detection of Pol V-dependent transcripts from AtSN1-B and IGN5. TUB8 was used as an internal control, and the RNA samples without reverse transcription (No RT) were used as negative control, indicating no DNA contamination.
Figure 4.
Developmental phenotypes of rdm4 mutant plants and mutant complementation. (A) The pleiotropic developmental phenotypes of ros1rdm4 and rdm4 mutant plants. (Panels I) ros1 plants. (Panels II) ros1rdm4 plants. (B) Diagram of the RDM4 gene showing the position of exons (boxes), introns (lines), and site of T-DNA insertion. (C) The RDM4 genomic construct complements the luminescence phenotype of ros1rdm4 in a representative T2 transgenic line. (D) The RDM4 transgene complements the developmental defects of ros1rdm4. The complemented plants shown are T1 transgenic lines.
Figure 5.
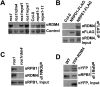
RDM4 interacts with NRPE1 and RPB1. (A) The RDM4 protein level was assessed by Western blot analysis in the indicated genotypes. A nonspecific band (Control) is shown as loading control. (B) Coimmunoprecipitation between RDM4 and NRPE1-Flag. The protein extracts from the indicated genotypes were precipitated by anti-Flag antibody-conjugated beads, and the precipitates were detected by anti-RDM4 and anti-Flag antibodies. (C) Coimmunoprecipitation between RDM4 and RPB1. The protein extracts from ros1 and ros1rdm4 were precipitated by anti-RDM4 antibody-conjugated beads, and the precipitates were detected by anti-RPB1 and anti-RDM4 antibodies. (D) The protein extracts from wild-type and YFP-RDM4 transgenic plants were precipitated by anti-RPB1 antibody-conjugated beads, and the precipitates were detected by anti-YFP and anti-RPB1 antibodies.
Similar articles
- RNA-directed DNA methylation and plant development require an IWR1-type transcription factor.
Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, Lind M, Schmitt MJ, Simon SA, Gurazada SG, Meyers BC, Lorkovic ZJ, Matzke AJ, Matzke M. Kanno T, et al. EMBO Rep. 2010 Jan;11(1):65-71. doi: 10.1038/embor.2009.246. Epub 2009 Dec 11. EMBO Rep. 2010. PMID: 20010803 Free PMC article. - NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - RNA polymerases IV and V influence the 3' boundaries of Polymerase II transcription units in Arabidopsis.
McKinlay A, Podicheti R, Wendte JM, Cocklin R, Rusch DB. McKinlay A, et al. RNA Biol. 2018 Feb 1;15(2):269-279. doi: 10.1080/15476286.2017.1409930. Epub 2017 Dec 21. RNA Biol. 2018. PMID: 29199514 Free PMC article. - Molecular mechanisms of the RNA polymerases in plant RNA-directed DNA methylation.
Xie G, Du X, Hu H, Du J. Xie G, et al. Trends Biochem Sci. 2024 Mar;49(3):247-256. doi: 10.1016/j.tibs.2023.11.005. Epub 2023 Dec 9. Trends Biochem Sci. 2024. PMID: 38072749 Review. - RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants.
Matzke MA, Kanno T, Matzke AJ. Matzke MA, et al. Annu Rev Plant Biol. 2015;66:243-67. doi: 10.1146/annurev-arplant-043014-114633. Epub 2014 Dec 10. Annu Rev Plant Biol. 2015. PMID: 25494460 Review.
Cited by
- slc7a6os gene plays a critical role in defined areas of the developing CNS in zebrafish.
Benini A, Cignarella F, Calvarini L, Mantovanelli S, Giacopuzzi E, Zizioli D, Borsani G. Benini A, et al. PLoS One. 2015 Mar 24;10(3):e0119696. doi: 10.1371/journal.pone.0119696. eCollection 2015. PLoS One. 2015. PMID: 25803583 Free PMC article. - Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing.
Haag JR, Pikaard CS. Haag JR, et al. Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):483-92. doi: 10.1038/nrm3152. Nat Rev Mol Cell Biol. 2011. PMID: 21779025 Review. - The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci.
Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. Liu ZW, et al. PLoS Genet. 2014 Jan;10(1):e1003948. doi: 10.1371/journal.pgen.1003948. Epub 2014 Jan 22. PLoS Genet. 2014. PMID: 24465213 Free PMC article. - A conserved Pol II elongator SPT6L mediates Pol V transcription to regulate RNA-directed DNA methylation in Arabidopsis.
Liu Y, Shu J, Zhang Z, Ding N, Liu J, Liu J, Cui Y, Wang C, Chen C. Liu Y, et al. Nat Commun. 2024 May 25;15(1):4460. doi: 10.1038/s41467-024-48940-8. Nat Commun. 2024. PMID: 38796517 Free PMC article. - A Role for MINIYO and QUATRE-QUART2 in the Assembly of RNA Polymerases II, IV, and V in Arabidopsis.
Li Y, Yuan Y, Fang X, Lu X, Lian B, Zhao G, Qi Y. Li Y, et al. Plant Cell. 2018 Feb;30(2):466-480. doi: 10.1105/tpc.17.00380. Epub 2018 Jan 18. Plant Cell. 2018. PMID: 29352065 Free PMC article.
References
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. - PubMed
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. - PubMed
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM059138/GM/NIGMS NIH HHS/United States
- R01 GM070795/GM/NIGMS NIH HHS/United States
- R01GM059138/GM/NIGMS NIH HHS/United States
- R01GM070795/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases