Improvements to services at the European Nucleotide Archive - PubMed (original) (raw)
. 2010 Jan;38(Database issue):D39-45.
doi: 10.1093/nar/gkp998. Epub 2009 Nov 11.
Ruth Akhtar, Ewan Birney, James Bonfield, Lawrence Bower, Matt Corbett, Ying Cheng, Fehmi Demiralp, Nadeem Faruque, Neil Goodgame, Richard Gibson, Gemma Hoad, Christopher Hunter, Mikyung Jang, Steven Leonard, Quan Lin, Rodrigo Lopez, Michael Maguire, Hamish McWilliam, Sheila Plaister, Rajesh Radhakrishnan, Siamak Sobhany, Guy Slater, Petra Ten Hoopen, Franck Valentin, Robert Vaughan, Vadim Zalunin, Daniel Zerbino, Guy Cochrane
Affiliations
- PMID: 19906712
- PMCID: PMC2808951
- DOI: 10.1093/nar/gkp998
Improvements to services at the European Nucleotide Archive
Rasko Leinonen et al. Nucleic Acids Res. 2010 Jan.
Abstract
The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) is Europe's primary nucleotide sequence archival resource, safeguarding open nucleotide data access, engaging in worldwide collaborative data exchange and integrating with the scientific publication process. ENA has made significant contributions to the collaborative nucleotide archival arena as an active proponent of extending the traditional collaboration to cover capillary and next-generation sequencing information. We have continued to co-develop data and metadata representation formats with our collaborators for both data exchange and public data dissemination. In addition to the DDBJ/EMBL/GenBank feature table format, we share metadata formats for capillary and next-generation sequencing traces and are using and contributing to the NCBI SRA Toolkit for the long-term storage of the next-generation sequence traces. During the course of 2009, ENA has significantly improved sequence submission, search and access functionalities provided at EMBL-EBI. In this article, we briefly describe the content and scope of our archive and introduce major improvements to our services.
Figures
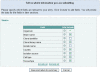
Figure 1.
Selection of the sequence submission type.
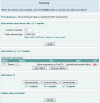
Figure 2.
Selection of the fields to include in the submission.
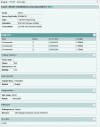
Figure 3.
Submission summary page.
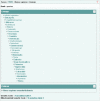
Figure 4.
ENA Browser project page.
Figure 5.
ENA Browser taxonomy page.
Similar articles
- Archiving next generation sequencing data.
Shumway M, Cochrane G, Sugawara H. Shumway M, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D870-1. doi: 10.1093/nar/gkp1078. Epub 2009 Dec 3. Nucleic Acids Res. 2010. PMID: 19965774 Free PMC article. - The European Nucleotide Archive.
Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tárraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, Hoad G, Jang M, Pakseresht N, Plaister S, Radhakrishnan R, Reddy K, Sobhany S, Ten Hoopen P, Vaughan R, Zalunin V, Cochrane G. Leinonen R, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D28-31. doi: 10.1093/nar/gkq967. Epub 2010 Oct 23. Nucleic Acids Res. 2011. PMID: 20972220 Free PMC article. - The Sequence Read Archive: explosive growth of sequencing data.
Kodama Y, Shumway M, Leinonen R; International Nucleotide Sequence Database Collaboration. Kodama Y, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D54-6. doi: 10.1093/nar/gkr854. Epub 2011 Oct 18. Nucleic Acids Res. 2012. PMID: 22009675 Free PMC article. - EMBL's European Bioinformatics Institute (EMBL-EBI) in 2023.
Thakur M, Buniello A, Brooksbank C, Gurwitz KT, Hall M, Hartley M, Hulcoop DG, Leach AR, Marques D, Martin M, Mithani A, McDonagh EM, Mutasa-Gottgens E, Ochoa D, Perez-Riverol Y, Stephenson J, Varadi M, Velankar S, Vizcaino JA, Witham R, McEntyre J. Thakur M, et al. Nucleic Acids Res. 2024 Jan 5;52(D1):D10-D17. doi: 10.1093/nar/gkad1088. Nucleic Acids Res. 2024. PMID: 38015445 Free PMC article. Review. - Efficient compression of SARS-CoV-2 genome data using Nucleotide Archival Format.
Kryukov K, Jin L, Nakagawa S. Kryukov K, et al. Patterns (N Y). 2022 Sep 9;3(9):100562. doi: 10.1016/j.patter.2022.100562. Epub 2022 Jul 7. Patterns (N Y). 2022. PMID: 35818472 Free PMC article. Review.
Cited by
- YACHT: an ANI-based statistical test to detect microbial presence/absence in a metagenomic sample.
Koslicki D, White S, Ma C, Novikov A. Koslicki D, et al. Bioinformatics. 2024 Feb 1;40(2):btae047. doi: 10.1093/bioinformatics/btae047. Bioinformatics. 2024. PMID: 38268451 Free PMC article. - The European Bioinformatics Institute's data resources.
Brooksbank C, Cameron G, Thornton J. Brooksbank C, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D17-25. doi: 10.1093/nar/gkp986. Epub 2009 Nov 24. Nucleic Acids Res. 2010. PMID: 19934258 Free PMC article. - UniProt Knowledgebase: a hub of integrated protein data.
Magrane M; UniProt Consortium. Magrane M, et al. Database (Oxford). 2011 Mar 29;2011:bar009. doi: 10.1093/database/bar009. Print 2011. Database (Oxford). 2011. PMID: 21447597 Free PMC article. - Human and chicken TLR pathways: manual curation and computer-based orthology analysis.
Gillespie M, Shamovsky V, D'Eustachio P. Gillespie M, et al. Mamm Genome. 2011 Feb;22(1-2):130-8. doi: 10.1007/s00335-010-9296-0. Epub 2010 Oct 30. Mamm Genome. 2011. PMID: 21052677 Free PMC article. - SkewC: Identifying cells with skewed gene body coverage in single-cell RNA sequencing data.
Abugessaisa I, Hasegawa A, Noguchi S, Cardon M, Watanabe K, Takahashi M, Suzuki H, Katayama S, Kere J, Kasukawa T. Abugessaisa I, et al. iScience. 2022 Jan 15;25(2):103777. doi: 10.1016/j.isci.2022.103777. eCollection 2022 Feb 18. iScience. 2022. PMID: 35146392 Free PMC article.
References
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. - PubMed
- Ansorge WJ. Next-generation DNA sequencing techniques. N. Biotechnol. 2009;25:195–203. - PubMed