Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells - PubMed (original) (raw)
Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells
Xue-Jun Li et al. Development. 2009 Dec.
Abstract
The directed differentiation of forebrain neuronal types from human embryonic stem cells (hESCs) has not been achieved. Here, we show that hESCs differentiate to telencephalic progenitors with a predominantly dorsal identity in a chemically defined medium without known morphogens. This is attributed to endogenous Wnt signaling, which upregulates the truncated form of GLI3, a repressor of sonic hedgehog (SHH). A high concentration of SHH, or the inhibition of Wnt by dickkopf 1 (DKK1) together with a low concentration of SHH, almost completely converts the primitive dorsal precursors to ventral progenitors, which is partially achieved through both downregulation of the truncated GLI3 and upregulation of full-length GLI3 expression. These dorsal and ventral telencephalic progenitors differentiate to functional glutamatergic and GABAergic neurons, respectively. Thus, although hESCs generate dorsal telencephalic cells, as opposed to ventral progenitors in other vertebrates, in the absence of exogenous morphogens, human cells use a similar molecular mechanism to control the dorsal versus ventral fate. The coordination of Wnt and SHH signaling through GLI3 represents a novel mechanism that regulates ventral-dorsal patterning in the development of forebrain neuronal subtypes.
Figures
Fig. 1.
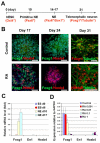
Generation of telencephalic neurons from hESCs. (A) A schematic of the procedure for generating telencephalic progenitors from hESCs. (B) In the control group, the expression of FOXG1 was maintained at 1 month after differentiation, whereas retinoic acid treatment (RA; 0.1 μM) resulted in the loss of FOXG1 expression and the generation of HOXB4+ cells. (C) The change in expression of anteroposterior homeodomain genes during neural differentiation using qPCR. ES, embryonic stem; EB, embryoid body; NE, neuroepithelium. (D) RA, added to the primitive neuroepithelial (NE) cells (at day 10) for 1 week, induced a dose-dependent repression of FOXG1 and expression of HOXB4 mRNA. Data are presented as mean±s.d., _n_=3. Blue, Hoechst-stained nuclei. Scale bars: 50 μm.
Fig. 2.
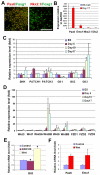
Neuroepithelial cells display a dorsal identity. (A) At 1 month after differentiation in the absence of exogenous morphogens, the majority of cells were positive for both FOXG1 and PAX6, whereas almost no cells were NKX2-1+. (B) qPCR showed the high expression level of PAX6 and EMX1 mRNA, but low level of NKX2-1 and DLX2 mRNA by NE cells at day 17. (C) Microarray analysis indicated little change in SHH signaling components except a significant increase in GLI3. (D) There is an increase in Wnt proteins and frizzled (FZD) proteins after day 6 of differentiation. (E) WNT3A-conditioned medium upregulated, whereas DKK1 (100 ng/ml, from day 10 to day 17) decreased, GLI3 expression at the mRNA level as revealed by qPCR. (F) Addition of WNT3A-conditioned medium from day 10 to day 17 resulted in an increase in the expression of PAX6 and EMX1 mRNA. Data are presented as mean±s.d. *, P<0.05 versus the control group, _n_=4. Scale bar: 50 μm.
Fig. 3.
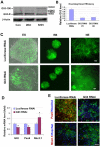
Role of GLI3 in dorsal-ventral pattering. (A) Western blotting showed the expression of GLI3 proteins by NE cells from control, WNT3A-treated and SHH (500 ng/ml)-treated groups. GLI3-190, full-length active form; GLI3-R, truncated repressive form. (B) HEK293 cells were transiently transfected with vectors containing luciferase shRNA or GLI3 shRNA. qRT-PCR analysis showed a significant decrease in GLI3 mRNA expression in the GLI3 RNAi, but not the luciferase RNAi, group. Data are presented as mean±s.d., _n_=3. Two sets of shRNAs possess similar knockdown efficiency. (C) Human ESCs infected with lentiviruses for luciferase RNAi and GLI3 RNAi showed a similar population of RNAi-expressing cells, as indicated by GFP expression in the differentiated NE cells. Note that not all cells were GFP+ as the transduction efficiency was not 100%. (D) qRT-PCR showing the expression of GLI3, PAX6, and NKX2-1 by NE cells at day 17. Data are presented as mean±s.d., _n_=3. *, P<0.05 between GLI3 RNAi and luciferase RNAi groups by two-sided _t_-test. (E) The expression of PAX6 and NKX2-1 proteins in cells from both luciferase RNAi and GLI3 RNAi groups 1 month after differentiation. Blue, Hoechst-stained nuclei. Scale bars: 50 μm in C; 20 μm in E.
Fig. 4.
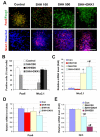
Specification of dorsal and ventral telencephalic progenitors. (A,B) Primitive neuroepithelial (NE) cells (day 10) were treated with SHH and/or DKK1 and the populations of NKX2-1+ and PAX6+ cells were compared at day 28. A high concentration of SHH, or SHH and DKK1 together, converted PAX6-expressing cells to NKX2-1+ cells. (C-E) The expression of NKX2-1 (C), PAX6 (D) and GLI3 (E) was measured by qRT-PCR 7 days after the primitive NE cells were cultured in basic medium (control) or in the presence of SHH (100 or 500 ng/ml), DKK1 (100 ng/ml), or DKK1 (100 ng/ml) plus SHH (100 ng/ml). Data are presented as mean±s.d. *, P<0.05 versus the control group, #, P<0.05 versus the SHH-treated (100 ng/ml) group, _n_=4. Blue, Hoechst-stained nuclei. Scale bar: 50 μm.
Fig. 5.
Differentiation of glutamatergic and GABAergic neurons from dorsal and ventral progenitors. (A,B) Comparison of the expression of TBR1, CTIP2 and ISLET1 between the neurons that were developed from dorsal (control group) and ventral (DKK1 plus SHH group) progenitors. (C-E) Dorsal progenitors differentiated into cells that were positive for TBR1 (C), CTIP2 (D) and VGLUT1 (E). (F-H) Ventral progenitors differentiated into neurons that expressed GAD (F), GABA (G) and DARPP32 (H). (I) Western blot analysis showing differential expression of TBR1, DARPP32 and GAD65/67 between cultures patterned for dorsal and ventral identities. Blue, Hoechst-stained nuclei. Scale bars: 50 μm.
Fig. 6.
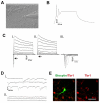
Human ESC-derived neurons functionally mature in vitro. (A) A phase-contrast image of a neuronal culture during patch clamp recordings. (B) Action potentials are elicited upon a 20 pA current injection in neurons differentiated for 12 weeks in vitro. (C) Inward and outward currents are elicited upon voltage steps (−50 mV to 50 mV) in a 12-week-old neuron (i). Application of 1 μM tetrodotoxin (ii) completely eliminated inward currents (i,ii, arrows) that are characteristic of Na+ currents (iii). (D) In the ventrally patterned neuronal population, spontaneous postsynaptic currents were recorded from 10-week-old neurons (i) and application of 50 μM bicucculine completely inhibited postsynaptic currents (ii). (E) Immunostaining showed the expression of TBR1 by a biocytin-labeled neuron after patch clamping. Scale bar: 50 μm.
Similar articles
- Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals.
Danjo T, Eiraku M, Muguruma K, Watanabe K, Kawada M, Yanagawa Y, Rubenstein JL, Sasai Y. Danjo T, et al. J Neurosci. 2011 Feb 2;31(5):1919-33. doi: 10.1523/JNEUROSCI.5128-10.2011. J Neurosci. 2011. PMID: 21289201 Free PMC article. - Patterning the dorsal telencephalon: a role for sonic hedgehog?
Rash BG, Grove EA. Rash BG, et al. J Neurosci. 2007 Oct 24;27(43):11595-603. doi: 10.1523/JNEUROSCI.3204-07.2007. J Neurosci. 2007. PMID: 17959802 Free PMC article. - Ectopic sonic hedgehog signaling impairs telencephalic dorsal midline development: implication for human holoprosencephaly.
Huang X, Litingtung Y, Chiang C. Huang X, et al. Hum Mol Genet. 2007 Jun 15;16(12):1454-68. doi: 10.1093/hmg/ddm096. Epub 2007 Apr 27. Hum Mol Genet. 2007. PMID: 17468181 - New perspectives on the mechanisms establishing the dorsal-ventral axis of the spinal cord.
Andrews MG, Kong J, Novitch BG, Butler SJ. Andrews MG, et al. Curr Top Dev Biol. 2019;132:417-450. doi: 10.1016/bs.ctdb.2018.12.010. Epub 2018 Dec 26. Curr Top Dev Biol. 2019. PMID: 30797516 Free PMC article. Review. - Signaling cascade coordinating growth of dorsal and ventral tissues of the vertebrate brain, with special reference to the involvement of Sonic Hedgehog signaling.
Ishibashi M, Saitsu H, Komada M, Shiota K. Ishibashi M, et al. Anat Sci Int. 2005 Mar;80(1):30-6. doi: 10.1111/j.1447-073x.2005.00096.x. Anat Sci Int. 2005. PMID: 15794128 Review.
Cited by
- Human Stem Cell-Derived Astrocytes: Specification and Relevance for Neurological Disorders.
Tyzack G, Lakatos A, Patani R. Tyzack G, et al. Curr Stem Cell Rep. 2016;2(3):236-247. doi: 10.1007/s40778-016-0049-1. Epub 2016 Jun 3. Curr Stem Cell Rep. 2016. PMID: 27547709 Free PMC article. Review. - Modeling simple repeat expansion diseases with iPSC technology.
Jaworska E, Kozlowska E, Switonski PM, Krzyzosiak WJ. Jaworska E, et al. Cell Mol Life Sci. 2016 Nov;73(21):4085-100. doi: 10.1007/s00018-016-2284-0. Epub 2016 Jun 3. Cell Mol Life Sci. 2016. PMID: 27261369 Free PMC article. Review. - Pluripotent Stem Cells for Brain Repair: Protocols and Preclinical Applications in Cortical and Hippocampal Pathologies.
Alia C, Terrigno M, Busti I, Cremisi F, Caleo M. Alia C, et al. Front Neurosci. 2019 Aug 6;13:684. doi: 10.3389/fnins.2019.00684. eCollection 2019. Front Neurosci. 2019. PMID: 31447623 Free PMC article. Review. - WNT/NOTCH Pathway Is Essential for the Maintenance and Expansion of Human MGE Progenitors.
Ma L, Wang Y, Hui Y, Du Y, Chen Z, Feng H, Zhang S, Li N, Song J, Fang Y, Xu X, Shi L, Zhang B, Cheng J, Zhou S, Liu L, Zhang X. Ma L, et al. Stem Cell Reports. 2019 May 14;12(5):934-949. doi: 10.1016/j.stemcr.2019.04.007. Epub 2019 May 2. Stem Cell Reports. 2019. PMID: 31056478 Free PMC article. - Inhibiting mitochondrial fission rescues degeneration in hereditary spastic paraplegia neurons.
Chen Z, Chai E, Mou Y, Roda RH, Blackstone C, Li XJ. Chen Z, et al. Brain. 2022 Nov 21;145(11):4016-4031. doi: 10.1093/brain/awab488. Brain. 2022. PMID: 35026838 Free PMC article.
References
- Ahn S., Joyner A. L. (2004). Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505-516 - PubMed
- Alvarez-Medina R., Cayuso J., Okubo T., Takada S., Marti E. (2008). Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development 135, 237-247 - PubMed
- Anderson K. D., Reiner A. (1991). Immunohistochemical localization of DARPP-32 in striatal projection neurons and striatal interneurons: implications for the localization of D1-like dopamine receptors on different types of striatal neurons. Brain Res. 568, 235-243 - PubMed
- Arlotta P., Molyneaux B. J., Chen J., Inoue J., Kominami R., Macklis J. D. (2005). Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207-221 - PubMed
- Bai C. B., Stephen D., Joyner A. L. (2004). All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6, 103-115 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 HD003352/HD/NICHD NIH HHS/United States
- R01 NS045926-06A2/NS/NINDS NIH HHS/United States
- R01 NS045926-05/NS/NINDS NIH HHS/United States
- T32 GM008692/GM/NIGMS NIH HHS/United States
- R01 NS045926/NS/NINDS NIH HHS/United States
- P30 HD003352-43/HD/NICHD NIH HHS/United States
- P30 HD03352/HD/NICHD NIH HHS/United States
- P30 HD003352-42/HD/NICHD NIH HHS/United States
- R01 NS045926-04/NS/NINDS NIH HHS/United States
- R21 NS055261/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources