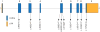Exome sequencing identifies the cause of a mendelian disorder - PubMed (original) (raw)
doi: 10.1038/ng.499. Epub 2009 Nov 13.
Kati J Buckingham, Choli Lee, Abigail W Bigham, Holly K Tabor, Karin M Dent, Chad D Huff, Paul T Shannon, Ethylin Wang Jabs, Deborah A Nickerson, Jay Shendure, Michael J Bamshad
Affiliations
- PMID: 19915526
- PMCID: PMC2847889
- DOI: 10.1038/ng.499
Exome sequencing identifies the cause of a mendelian disorder
Sarah B Ng et al. Nat Genet. 2010 Jan.
Abstract
We demonstrate the first successful application of exome sequencing to discover the gene for a rare mendelian disorder of unknown cause, Miller syndrome (MIM%263750). For four affected individuals in three independent kindreds, we captured and sequenced coding regions to a mean coverage of 40x and sufficient depth to call variants at approximately 97% of each targeted exome. Filtering against public SNP databases and eight HapMap exomes for genes with two previously unknown variants in each of the four individuals identified a single candidate gene, DHODH, which encodes a key enzyme in the pyrimidine de novo biosynthesis pathway. Sanger sequencing confirmed the presence of DHODH mutations in three additional families with Miller syndrome. Exome sequencing of a small number of unrelated affected individuals is a powerful, efficient strategy for identifying the genes underlying rare mendelian disorders and will likely transform the genetic analysis of monogenic traits.
Figures
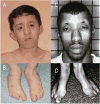
Figure 1. Clinical characteristics of an individual with Miller syndrome (A,B) and an individual with methotrexate embryopathy (C,D)
A 9 year-old boy with Miller syndrome (A and B) caused by mutations in DHODH. Facial anomalies (A) include cupped ears, coloboma of the lower eyelids, prominent nose, micrognathia and absence of the 5th digits of the feet (B). A 26 year-old man with methotrexate embryopathy (C and D). Note the cupped ears, hypertelorism, sparse eyebrows, and prominent nose (C) accompanied by absence of the 4th and 5th digits of the feet (D). C and D are reprinted with permission from Bawle et al. Teratology 57:51-55 (1978).
Figure 2. Genomic structure of the exons encoding the open reading frame of DHODH
DHODH is composed of 9 exons that encode untranslated regions (orange) and protein coding sequence (blue). Arrows indicate the locations of 11 different mutations found in 6 families with Miller syndrome.
Comment in
- Exome sequencing makes medical genomics a reality.
Biesecker LG. Biesecker LG. Nat Genet. 2010 Jan;42(1):13-4. doi: 10.1038/ng0110-13. Nat Genet. 2010. PMID: 20037612 - Exome sequencing: locating causative genes in rare disorders.
Pussegoda KA. Pussegoda KA. Clin Genet. 2010 Jul;78(1):32-3. doi: 10.1111/j.1399-0004.2010.01414_1.x. Clin Genet. 2010. PMID: 20597920 No abstract available. - Exome sequencing: expanding the genetic testing toolbox.
Kobelka CE. Kobelka CE. Clin Genet. 2010 Aug;78(2):132-4. doi: 10.1111/j.1399-0004.2010.01452_1.x. Clin Genet. 2010. PMID: 20662854 No abstract available.
Similar articles
- Exome sequencing makes medical genomics a reality.
Biesecker LG. Biesecker LG. Nat Genet. 2010 Jan;42(1):13-4. doi: 10.1038/ng0110-13. Nat Genet. 2010. PMID: 20037612 - Miller (Genee-Wiedemann) syndrome represents a clinically and biochemically distinct subgroup of postaxial acrofacial dysostosis associated with partial deficiency of DHODH.
Rainger J, Bengani H, Campbell L, Anderson E, Sokhi K, Lam W, Riess A, Ansari M, Smithson S, Lees M, Mercer C, McKenzie K, Lengfeld T, Gener Querol B, Branney P, McKay S, Morrison H, Medina B, Robertson M, Kohlhase J, Gordon C, Kirk J, Wieczorek D, Fitzpatrick DR. Rainger J, et al. Hum Mol Genet. 2012 Sep 15;21(18):3969-83. doi: 10.1093/hmg/dds218. Epub 2012 Jun 12. Hum Mol Genet. 2012. PMID: 22692683 - Revisiting Mendelian disorders through exome sequencing.
Ku CS, Naidoo N, Pawitan Y. Ku CS, et al. Hum Genet. 2011 Apr;129(4):351-70. doi: 10.1007/s00439-011-0964-2. Epub 2011 Feb 18. Hum Genet. 2011. PMID: 21331778 Review. - Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome.
Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Ng SB, et al. Nat Genet. 2010 Sep;42(9):790-3. doi: 10.1038/ng.646. Epub 2010 Aug 15. Nat Genet. 2010. PMID: 20711175 Free PMC article. - Protein production, kinetic and biophysical characterization of three human dihydroorotate dehydrogenase mutants associated with Miller syndrome.
Orozco Rodriguez JM, Krupinska E, Wacklin-Knecht H, Knecht W. Orozco Rodriguez JM, et al. Nucleosides Nucleotides Nucleic Acids. 2022;41(12):1318-1336. doi: 10.1080/15257770.2021.2023749. Epub 2022 Jan 30. Nucleosides Nucleotides Nucleic Acids. 2022. PMID: 35094635 Review.
Cited by
- A mild skeletal phenotype with overlapping features of Miller syndrome and functional characterisation of two new variants of human dihydroorotate dehydrogenase.
Mero IL, Orozco Rodriguez JM, Bjørgo K, Hankin RA, Krupinska E, Kulseth MA, Rossow MA, Knecht W. Mero IL, et al. Heliyon. 2024 Sep 27;10(19):e38659. doi: 10.1016/j.heliyon.2024.e38659. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39430512 Free PMC article. - Exploring the design of clinical research studies on the efficacy mechanisms in type 2 diabetes mellitus.
Guan H, Zhao S, Li J, Wang Y, Niu P, Zhang Y, Zhang Y, Fang X, Miao R, Tian J. Guan H, et al. Front Endocrinol (Lausanne). 2024 Sep 20;15:1363877. doi: 10.3389/fendo.2024.1363877. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39371930 Free PMC article. Review. - Federated analysis of autosomal recessive coding variants in 29,745 developmental disorder patients from diverse populations.
Chundru VK, Zhang Z, Walter K, Lindsay SJ, Danecek P, Eberhardt RY, Gardner EJ, Malawsky DS, Wigdor EM, Torene R, Retterer K, Wright CF, Ólafsdóttir H, Guillen Sacoto MJ, Ayaz A, Akbeyaz IH, Türkdoğan D, Al Balushi AI, Bertoli-Avella A, Bauer P, Szenker-Ravi E, Reversade B, McWalter K, Sheridan E, Firth HV, Hurles ME, Samocha KE, Ustach VD, Martin HC. Chundru VK, et al. Nat Genet. 2024 Oct;56(10):2046-2053. doi: 10.1038/s41588-024-01910-8. Epub 2024 Sep 23. Nat Genet. 2024. PMID: 39313616 Free PMC article. - An atypical form of 60S ribosomal subunit in Diamond-Blackfan anemia linked to RPL17 variants.
Fellmann F, Saunders C, O'Donohue MF, Reid DW, McFadden KA, Montel-Lehry N, Yu C, Fang M, Zhang J, Royer-Bertrand B, Farinelli P, Karboul N, Willer JR, Fievet L, Bhuiyan ZA, Kleinhenz AL, Jadeau J, Fulbright J, Rivolta C, Renella R, Katsanis N, Beckmann JS, Nicchitta CV, Da Costa L, Davis EE, Gleizes PE. Fellmann F, et al. JCI Insight. 2024 Aug 1;9(17):e172475. doi: 10.1172/jci.insight.172475. JCI Insight. 2024. PMID: 39088281 Free PMC article.
References
- Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotechnology. 2008;26:1135–1145. - PubMed
- Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL094976/HL/NHLBI NIH HHS/United States
- R01 HL094976-01/HL/NHLBI NIH HHS/United States
- R21 HG004749/HG/NHGRI NIH HHS/United States
- R21 HG004749-01/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous