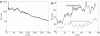Membrane-bending mechanism of amphiphysin N-BAR domains - PubMed (original) (raw)
Membrane-bending mechanism of amphiphysin N-BAR domains
Anton Arkhipov et al. Biophys J. 2009.
Abstract
BAR domains are highly conserved protein domains participating in a diversity of cellular processes that involve membrane remodeling. The mechanisms underlying such remodeling are debated. For the relatively well-studied case of amphiphysin N-BAR domain, one suggested mechanism involves scaffolding, i.e., binding of a negatively charged membrane to the protein's positively charged curved surface. An alternative mechanism suggests that insertion of the protein's N-terminal amphipathic segments (N-helices H0) into the membrane leads to bending. Here, we address the issue through all-atom and coarse-grained simulations of multiple amphiphysin N-BAR domains and their components interacting with a membrane. We observe that complete N-BAR domains and BAR domains without H0s bend the membrane, but H0s alone do not, which suggests that scaffolding, rather than helix insertion, plays a key role in membrane sculpting by amphiphysin N-BAR domains.
Figures
Figure 1
Amphiphysin N-BAR domain and lipid membrane. (a) The all-atom structure of the N-BAR domain, with charged residues highlighted in positive (blue) and negative (red). (b) The membrane composed of 70% DOPC (cyan) and 30% DOPS (pink) lipids. The SBCG models (c) of the N-BAR domains and (d) of the membrane; the SBCG beads of the N-BAR domains are colored according to their charge, on a linear scale from red at −2|e| to blue at 2|e|. The N-BAR domains in panels a and c are depicted as viewed from the side and from the top. The N-terminal segments H0 are highlighted by dashed ovals.
Figure 2
All-atom simulations of membrane bending by whole N-BAR domains and by their components. (a) Simulation NBAR-init (originally reported in (21)), (b) simulation H0, and (c) simulation noH0. The simulations are defined in Table 1. For each simulation, top and side views of the initial conformation, and the side view of the final conformation, are shown. In panel a, images of the initial conformation also include the water box, which is not shown in the remaining images.
Figure 3
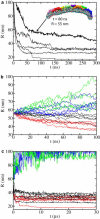
SBCG simulations of membrane bending by N-BAR domains and their components. (a) Simulation CG-NBAR-init. The conformation from this simulation at t = 80 ns is chosen as a starting point for other SBCG simulations, namely, (b) CG-H0, (c) CG-mem, (d) CG-NBAR, and (e) CG-noH0. See Table 1 for details. In each case, five SBCG simulations were performed, and the snapshots from each are shown at t = 30 ns (corresponding to ∼100 ns for all-atom simulation, see Fig. 2, b and c) and at t = 30 _μ_s. Initial conformations for each simulation are shown from the top and from the side.
Figure 4
Radius of membrane curvature, R, versus time, t, from all-atom simulations. (a) R for simulation NBAR-init (see Fig. 2_a_). (b) R values for simulations H0 and noH0 (see Fig. 2, b and c).
Figure 5
Radius of membrane curvature, R, versus time, t, in SBCG simulations. (a) Results from five SBCG simulations CG-NBAR-init (see also Fig. 4_a_). The thicker line is from the simulation that is most similar to the respective all-atom simulation (Fig. 2_a_). The arrow shows the moment at which the conformation is taken for the start of subsequent simulations CG-NBAR, CG-noH0, CG-H0, and CG-mem (see Fig. 3_a_). (b) Results from simulations CG-NBAR (black), CG-noH0 (red), CG-H0 (blue), and CG-mem (green) presented for the first 100 ns (see also Fig. 3, b_–_e, and Fig. 4_b_). (c) Results for times up to 30 _μ_s.
Figure 6
Dynamics of H0 segments in all-atom simulations. The arrangement of H0s at the beginning of simulation NBAR-init is shown at the top, the one by the end of simulation NBAR-init in the middle, and the one at the end of simulation H0 at the bottom. Only H0s are shown (e.g., BAR domains are present in simulation NBAR-init, but are not shown). Dimensions of the membrane are depicted schematically by a gray rectangle. All H0s are wrapped across the periodic cell boundaries, so that they all are viewed within the area of one periodic cell.
Similar articles
- Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations.
Blood PD, Voth GA. Blood PD, et al. Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15068-72. doi: 10.1073/pnas.0603917103. Epub 2006 Sep 28. Proc Natl Acad Sci U S A. 2006. PMID: 17008407 Free PMC article. - Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding.
Isas JM, Ambroso MR, Hegde PB, Langen J, Langen R. Isas JM, et al. Structure. 2015 May 5;23(5):873-881. doi: 10.1016/j.str.2015.02.014. Epub 2015 Apr 9. Structure. 2015. PMID: 25865245 Free PMC article. - BAR domains as sensors of membrane curvature: the amphiphysin BAR structure.
Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. Peter BJ, et al. Science. 2004 Jan 23;303(5657):495-9. doi: 10.1126/science.1092586. Epub 2003 Nov 26. Science. 2004. PMID: 14645856 - Attachment of rod-like (BAR) proteins and membrane shape.
Kabaso D, Gongadze E, Elter P, van Rienen U, Gimsa J, Kralj-Iglič V, Iglič A. Kabaso D, et al. Mini Rev Med Chem. 2011 Apr;11(4):272-82. doi: 10.2174/138955711795305353. Mini Rev Med Chem. 2011. PMID: 21428902 Free PMC article. Review. - BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature.
Itoh T, De Camilli P. Itoh T, et al. Biochim Biophys Acta. 2006 Aug;1761(8):897-912. doi: 10.1016/j.bbalip.2006.06.015. Epub 2006 Jul 28. Biochim Biophys Acta. 2006. PMID: 16938488 Review.
Cited by
- Capturing spontaneous partitioning of peripheral proteins using a biphasic membrane-mimetic model.
Arcario MJ, Ohkubo YZ, Tajkhorshid E. Arcario MJ, et al. J Phys Chem B. 2011 Jun 2;115(21):7029-37. doi: 10.1021/jp109631y. Epub 2011 May 11. J Phys Chem B. 2011. PMID: 21561114 Free PMC article. - Formation of protein-mediated bilayer tubes is governed by a snapthrough transition.
Mahapatra A, Rangamani P. Mahapatra A, et al. Soft Matter. 2023 Jun 14;19(23):4345-4359. doi: 10.1039/d2sm01676a. Soft Matter. 2023. PMID: 37255421 Free PMC article. - Physical basis of some membrane shaping mechanisms.
Simunovic M, Prévost C, Callan-Jones A, Bassereau P. Simunovic M, et al. Philos Trans A Math Phys Eng Sci. 2016 Jul 28;374(2072):20160034. doi: 10.1098/rsta.2016.0034. Philos Trans A Math Phys Eng Sci. 2016. PMID: 27298443 Free PMC article. Review. - The N-Terminal Amphipathic Helix of Endophilin Does Not Contribute to Its Molecular Curvature Generation Capacity.
Chen Z, Zhu C, Kuo CJ, Robustelli J, Baumgart T. Chen Z, et al. J Am Chem Soc. 2016 Nov 9;138(44):14616-14622. doi: 10.1021/jacs.6b06820. Epub 2016 Oct 28. J Am Chem Soc. 2016. PMID: 27755867 Free PMC article. - Counterion-mediated pattern formation in membranes containing anionic lipids.
Slochower DR, Wang YH, Tourdot RW, Radhakrishnan R, Janmey PA. Slochower DR, et al. Adv Colloid Interface Sci. 2014 Jun;208:177-88. doi: 10.1016/j.cis.2014.01.016. Epub 2014 Jan 30. Adv Colloid Interface Sci. 2014. PMID: 24556233 Free PMC article. Review.
References
- Marsh M., McMahon H.T. The structural era of endocytosis. Science. 1999;285:215–220. - PubMed
- Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. - PubMed
- Farsad K., Camilli P.D. Mechanisms of membrane deformation. Curr. Opin. Cell Biol. 2003;15:372–381. - PubMed
- McMahon H.T., Hills I.G. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 2004;16:379–391. - PubMed
- McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR005969/RR/NCRR NIH HHS/United States
- R01 GM067887/GM/NIGMS NIH HHS/United States
- R01-GM067887/GM/NIGMS NIH HHS/United States
- P41-RR005969/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases