Histone h3 exerts a key function in mitotic checkpoint control - PubMed (original) (raw)
Histone h3 exerts a key function in mitotic checkpoint control
Jianjun Luo et al. Mol Cell Biol. 2010 Jan.
Abstract
It has been firmly established that many interphase nuclear functions, including transcriptional regulation, are regulated by chromatin and histones. How mitotic progression and quality control might be influenced by histones is less well characterized. We show that histone H3 plays a crucial role in activating the spindle assembly checkpoint in response to a defect in mitosis. Prior to anaphase, all chromosomes must attach to spindles emanating from the opposite spindle pole bodies. The tension between sister chromatids generated by the poleward pulling force is an integral part of chromosome biorientation. Lack of tension due to erroneous attachment activates the spindle assembly checkpoint, which corrects the mistakes and ensures segregation fidelity. A histone H3 mutation impairs the ability of yeast cells to activate the checkpoint in a tensionless crisis, leading to missegregation and aneuploidy. The defects in tension sensing result directly from an attenuated H3-Sgo1p interaction essential for pericentric recruitment of Sgo1p. Reinstating the pericentric enrichment of Sgo1p alleviates the mitotic defects. Histone H3, and hence the chromatin, is thus a key factor transmitting the tension status to the spindle assembly checkpoint.
Figures
FIG. 1.
The G44S mutation confers pleiotropic phenotypes. (A) Yeast cells bearing the G44S allele as the sole copy of H3 were tested on YPD medium under the indicated conditions. Left panel, sixfold serially diluted log-phase cells were spotted for growth tests. All drug tests were conducted at 30°C. MMS, methyl methanesulfonate. (B) Position of G44 of H3 within a nucleosomal core particle. Left panels: two views of the crystal structure (Protein Data Bank entry 1ID3) based on White et al. (58). Right panels: closeup view of the 42Lys-Pro-Gly-Thr β turn (top) and the secondary structural domains (bottom) of histone H3. Numbers below the secondary structure are amino acid residues at the junctions of the indicated domains. The green dotted lines in the closeup represent possible hydrogen bonds between the carbonyl oxygen of K42 and the amide groups of G44 and Thr45. Hydrogen is not included. DNA is omitted for clarity.
FIG. 2.
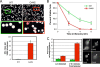
The G44S mutation causes chromosome instability. (A) Mitotic chromosome stability tests. Representative pictures of colonies on YPD plates are shown at the top. The bar graph was prepared from three independent experiments with the standard error of the mean. Colonies with an at least 50% continuous red sector were counted as the first-division chromosome (Chr.) loss. Colonies that were totally red, as a result of chromosome loss prior to cell plating on YPD, were excluded. (B) G44S mutant cells lose viability faster after short benomyl exposure. Log-phase cells were treated with 60 μg/ml benomyl for the indicated time, washed, counted, and spread onto benomyl-free YPD plates. Percent viability was calculated by dividing the total number of colonies by the number of cells inoculated (counted microscopically) and was normalized to that of the _T_0′ samples. Results are from at least three independent experiments. (C) Higher missegregation is associated with the G44S mutation. WT and G44S mutant cells with the TRP1 locus marked by GFP were treated with 30 μg/ml benomyl for 2 h, collected, and regrown in YPD medium containing α-factor for 2 h before fixation for microscopy. At least 200 unbudded cells with GFP dots were scored in four independent cell cultures of each strain. Green and red bars represent WT and G44S mutant cells, respectively. Error bars show standard deviations. Randomly selected images of two-dotted WT and G44S mutant cells (marked by white triangles) are shown on the right. w/o, without.
FIG. 3.
G44S mutant cells activate the spindle checkpoint in response to benomyl toxicity. (A) Comparable budding indices were obtained from WT and mutant cells. Benomyl (60 μg/ml) was added to log-phase cells. The percentage of large-budded cells (with daughter cells at least half the diameter of their mothers) was determined microscopically. (B) Pds1p was activated and stabilized in both normal and G44S mutant cells in the presence of benomyl. Comparable numbers of α-factor-arrested G1 cells were released at _T_0′ into YPD medium containing 0 or 30 μg/ml benomyl. The same volume of cell suspension was taken at the indicated times for boiling and whole-cell extract preparation, followed by anti-Myc Western blotting to quantify the abundance of Pds1p.
FIG. 4.
The G44S mutation impairs the tension-sensing function. MCD1 was placed under the control of the GAL1 promoter for glucose repression. The abundance of Myc-tagged Pds1p was analyzed by Western blotting in the presence (Gal to Gal) or absence (Gal to Glc) of Mcd1p. Experimental schemes are shown above the Western blot assay results. G-6-PDH, glucose-6-phosphate dehydrogenase.
FIG. 5.
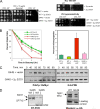
G44S mutant mitotic phenotypes are suppressed by overexpressing Sgo1p. (A) Sgo1p overproduction specifically rescues the mitotic phenotypes of G44S mutant cells. The SGO1 ORF was cloned into a 2μm plasmid bearing the promoter and terminator sequences of ADH1. WT and G44S mutant cells transformed with SGO1 or the corresponding vector were tested under the indicated conditions. (B) Left panel, cell viability test after benomyl treatment. This plot was generated from three independent experiments, and the error bars depict the standard deviations. Right panel, chromosome missegregation assay. Shown are percentages of two-GFP-dotted G1-phase cells. Data were collected from three or four independent cultures, and error bars represent standard deviations. See the legend to Fig. 2B and C for details. (C) Tension-sensing defects conferred by the G44S mutation are alleviated by SGO1 overexpression. G44S mutant cells receiving a 2μm plasmid with or without the _ADH1_-driven SGO1 gene were tested for the molecular response following MCD1 shutdown. Experiments were done exactly as those shown in Fig. 4. (D) Neither SGO1 transcription nor protein abundance is affected by the G44S mutation. Left, reverse transcription (RT)-PCR shows normal expression of SGO1 in the WT and G44S mutant strains. Right, Western blotting of C′-6×HA-tagged Sgo1p demonstrates the equal abundance of Sgo1p in these two strains. The loading control is a cross-reacting band from both yeast lysates.
FIG. 6.
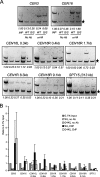
The G44S mutation selectively downregulates the pericentric recruitment of Sgo1p in vivo. (A) ChIP analysis of HA-tagged Sgo1p. Samples in all of the panels are arranged in the same order. The common internal control (marked by the star on the right) for multiplex PCRs is from within the ORF of the DED1 gene 386.2 kb to the right of CEN15. Targets of the PCR fragments and their distance to the cognate centromeres are listed at the top of each gel image. R, right; L, left. SPT15 is 313.3 kb to the right of CEN5. (B) Quantification of ChIP results. Ethidium bromide-stained DNA gel images were quantified by NIH Image. The intensity of each CEN or pericentric fragment was compared to that of the DED1 internal control (star). The ratio was then normalized to 0.1% input (INP) DNA (set at 1.0). Error bars represent standard deviations from at least three independent cell cultures for ChIP. Ab, antibody.
FIG. 7.
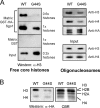
Physical interaction between H3 and Sgo1p is attenuated by the G44S mutation. (A) Pulldown assays assessing the interactions between bacterially expressed GST-3×HA-Sgo1p and histones and oligonucleosomes purified from WT or G44S mutant yeast cells. Trapped H3 was quantified by anti-H3 antibodies. (B) Far-Western assays detecting direct binding between H3 and Sgo1p. Yeast core histones were resolved, blotted onto a PVDF membrane, and incubated with GST-3×HA-Sgo1p. Sgo1p, trapped via association with H3, was probed by anti-HA antibodies. A parallel gel was stained with Coomassie blue R250 (CBR) to reveal the relative mobility of yeast histones.
FIG. 8.
Reestablishing pericentric Sgo1p recruitment by BD fusion. (A) ChIP assays assessing the distribution of Sgo1p and Sgo1p-BD at selective loci. G44S _sgo1_Δ mutant cells were transformed with an ARS CEN plasmid containing 3×HA-Sgo1p or 3×HA-Sgo1p-BD genes and analyzed by anti-HA ChIP. The assay conditions were identical to those described in the legend to Fig. 6. A nonspecific PCR fragment (N) amplified along with CEN16L 0.3 kb is marked with an arrow. (B) Comparable expression of 3×HA-Sgo1p and 3×HA-Sgo1p-BD. G44S _sgo1_Δ mutant cells bearing the indicated recombinant SGO1 construct were examined by anti-HA Western blotting. Equal loading was evidenced by Coomassie blue R250 staining (not shown) and by proteins that cross-reacted with the anti-HA antibodies. (C) Quantification of the ChIP results was done as detailed in the legend to Fig. 6B.
FIG. 9.
BD fusion partially rescues the tension-sensing defects of G44S mutant cells. (A) The indicated yeast strains expressing Sgo1p with or without the BD were tested for resistance to benomyl (A), for viability after benomyl exposure (B), and for the Pds1p level after MCD1 shutdown. See the legends to Fig. 2 and 4 for details. In panels B and C, only data from G44S mutant cells are shown.
Similar articles
- Critical roles of Shugoshin and histones as tension sensors during mitosis.
Buehl CJ, Kuo MH. Buehl CJ, et al. Curr Genet. 2018 Dec;64(6):1215-1219. doi: 10.1007/s00294-018-0846-4. Epub 2018 May 23. Curr Genet. 2018. PMID: 29796904 Review. - Tripartite Chromatin Localization of Budding Yeast Shugoshin Involves Higher-Ordered Architecture of Mitotic Chromosomes.
Deng X, Kuo MH. Deng X, et al. G3 (Bethesda). 2018 Aug 30;8(9):2901-2911. doi: 10.1534/g3.118.200522. G3 (Bethesda). 2018. PMID: 30002083 Free PMC article. - Identification of Tension Sensing Motif of Histone H3 in Saccharomyces cerevisiae and Its Regulation by Histone Modifying Enzymes.
Luo J, Deng X, Buehl C, Xu X, Kuo MH. Luo J, et al. Genetics. 2016 Nov;204(3):1029-1043. doi: 10.1534/genetics.116.192443. Epub 2016 Sep 26. Genetics. 2016. PMID: 27672091 Free PMC article. - A Failsafe for Sensing Chromatid Tension in Mitosis with the Histone H3 Tail in Saccharomyces cerevisiae.
Buehl CJ, Deng X, Luo J, Buranasudja V, Hazbun T, Kuo MH. Buehl CJ, et al. Genetics. 2018 Feb;208(2):565-578. doi: 10.1534/genetics.117.300606. Epub 2017 Dec 14. Genetics. 2018. PMID: 29242290 Free PMC article. - Monitoring the fidelity of mitotic chromosome segregation by the spindle assembly checkpoint.
Silva P, Barbosa J, Nascimento AV, Faria J, Reis R, Bousbaa H. Silva P, et al. Cell Prolif. 2011 Oct;44(5):391-400. doi: 10.1111/j.1365-2184.2011.00767.x. Cell Prolif. 2011. PMID: 21951282 Free PMC article. Review.
Cited by
- Diffuse Intrinsic Pontine Glioma: Molecular Landscape, Evolving Treatment Strategies and Emerging Clinical Trials.
Damodharan S, Lara-Velazquez M, Williamsen BC, Helgager J, Dey M. Damodharan S, et al. J Pers Med. 2022 May 20;12(5):840. doi: 10.3390/jpm12050840. J Pers Med. 2022. PMID: 35629262 Free PMC article. Review. - Characterization of hemocytes and hematopoietic cells of a freshwater crayfish based on single-cell transcriptome analysis.
Söderhäll I, Fasterius E, Ekblom C, Söderhäll K. Söderhäll I, et al. iScience. 2022 Aug 2;25(8):104850. doi: 10.1016/j.isci.2022.104850. eCollection 2022 Aug 19. iScience. 2022. PMID: 35996577 Free PMC article. - Critical roles of Shugoshin and histones as tension sensors during mitosis.
Buehl CJ, Kuo MH. Buehl CJ, et al. Curr Genet. 2018 Dec;64(6):1215-1219. doi: 10.1007/s00294-018-0846-4. Epub 2018 May 23. Curr Genet. 2018. PMID: 29796904 Review. - Tripartite Chromatin Localization of Budding Yeast Shugoshin Involves Higher-Ordered Architecture of Mitotic Chromosomes.
Deng X, Kuo MH. Deng X, et al. G3 (Bethesda). 2018 Aug 30;8(9):2901-2911. doi: 10.1534/g3.118.200522. G3 (Bethesda). 2018. PMID: 30002083 Free PMC article. - The dependence of shugoshin on Bub1-kinase activity is dispensable for the maintenance of spindle assembly checkpoint response in Cryptococcus neoformans.
Polisetty SD, Bhat K, Das K, Clark I, Hardwick KG, Sanyal K. Polisetty SD, et al. PLoS Genet. 2025 Jan 13;21(1):e1011552. doi: 10.1371/journal.pgen.1011552. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39804939 Free PMC article.
References
- Ahn, S. H., W. L. Cheung, J. Y. Hsu, R. L. Diaz, M. M. Smith, and C. D. Allis. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120:25-36. - PubMed
- Broder, Y. C., S. Katz, and A. Aronheim. 1998. The ras recruitment system, a novel approach to the study of protein-protein interactions. Curr. Biol. 8:1121-1124. - PubMed
- Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases