Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells - PubMed (original) (raw)
Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells
Sven Klunker et al. J Exp Med. 2009.
Abstract
Forkhead box P3 (FOXP3)(+)CD4(+)CD25(+) inducible regulatory T (iT reg) cells play an important role in immune tolerance and homeostasis. In this study, we show that the transforming growth factor-beta (TGF-beta) induces the expression of the Runt-related transcription factors RUNX1 and RUNX3 in CD4(+) T cells. This induction seems to be a prerequisite for the binding of RUNX1 and RUNX3 to three putative RUNX binding sites in the FOXP3 promoter. Inactivation of the gene encoding RUNX cofactor core-binding factor-beta (CBFbeta) in mice and small interfering RNA (siRNA)-mediated suppression of RUNX1 and RUNX3 in human T cells resulted in reduced expression of Foxp3. The in vivo conversion of naive CD4(+) T cells into Foxp3(+) iT reg cells was significantly decreased in adoptively transferred Cbfb(F/F) CD4-cre naive T cells into Rag2(-/-) mice. Both RUNX1 and RUNX3 siRNA silenced human T reg cells and Cbfb(F/F) CD4-cre mouse T reg cells showed diminished suppressive function in vitro. Circulating human CD4(+) CD25(high) CD127(-) T reg cells significantly expressed higher levels of RUNX3, FOXP3, and TGF-beta mRNA compared with CD4(+)CD25(-) cells. Furthermore, FOXP3 and RUNX3 were colocalized in human tonsil T reg cells. These data demonstrate Runx transcription factors as a molecular link in TGF-beta-induced Foxp3 expression in iT reg cell differentiation and function.
Figures
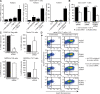
Figure 1.
RUNX1 and RUNX3 are involved in the induction of Foxp3 in iT reg cells. (A) RUNX1, RUNX3, and FOXP3 mRNA induction in human naive CD4+ T cells after alone or combined anti-CD2/3/28 mAb and TGF-β stimulation in the presence of IL-2. Real-time PCR of human naive CD4+ T cells after 48 h of culture. Bars show the mean ± SE of three independent experiments. (B) FOXP3 mRNA induction by anti-CD2/3/28 mAb and TGF-β in human naive CD4+ T cells is reduced after siRNA-mediated RUNX1/3 knockdown. Real-time PCR of RNA from human naive CD4+ T cells, transfected with RUNX1 and/or RUNX3 siRNA or with a control siRNA and cultured with anti-CD2/3/28, TGF-β and IL-2. Bars show the mean ± SD of three independent experiments. (C) FOXP3 mRNA is down-regulated in iT reg cells after siRNA-mediated knockdown of RUNX1 and RUNX3. Real-time PCR for FOXP3, T-bet, GATA3, and RORC2 from human naive CD4+ T cells, transfected with RUNX1 and RUNX3 siRNA or with scrambled siRNA (control) and cultured under iT reg, Th1, Th2, or Th17-driving conditions for 12 d. Bars show the mean ± SD of three independent experiments. (D) FOXP3 protein induction in iT reg cells is reduced after siRNA-mediated RUNX1/3 knockdown. Human naive CD4+ T cells were transfected with RUNX1 and/or RUNX3 siRNA or with a control siRNA and stimulated with anti-CD2/3/28 and TGF-β in the presence of IL-2. CD4 and intracellular FOXP3 analysis by flow cytometry after 72 h. One of three independent experiments is shown. Statistical differences were verified by the paired Student's t test. *, P < 0.05; **, P < 0.01.
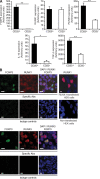
Figure 2.
RUNX1 and RUNX3 expression in Foxp3+ T reg cells and in human CD4+, CD127−**, CD25high cells.** (A) Real-time PCR analysis of CD4+, CD127−, and CD25high T reg cells and CD4+, CD127+, and CD25neg T cells isolated from human peripheral blood showed an increased expression of IL-10, TGF-β, FOXP3, and RUNX3 mRNA in CD25+ compared with CD25− cells. Bars show the mean ± SE of three independent experiments. (B) Human tonsil sections were analyzed by confocal microscopy. Tissue sections were stained for FOXP3, RUNX1, RUNX3, and DAPI or isotype controls. HEK cells RUNX1-transfected or not transfected served as additional control for RUNX1 staining. Data shown are representative from one of the three tissue samples with similar results. Bars, 5 µm. Statistical differences were verified by the paired Student's t test. *, P < 0.05; **, P < 0.01.
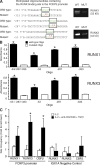
Figure 3.
Binding of RUNX1 and RUNX3 proteins to the predicted binding sites in the FOXP3 promoter. (A) Mutated and wild-type oligonucleotides are shown. The predicted RUNX binding sites are accentuated (boxed and in green letters) and stars mark mutations introduced into the binding site of the control oligonucleotides. Nuclear extracts from HEK293T cells were incubated with biotinylated oligonucleotides. The precipitated oligonucleotide–transcription factor complexes were separated by SDS-PAGE and identified by Western blotting with anti-RUNX1 and anti-RUNX3 antibodies. A mixture of all three oligonucleotides with the predicted binding sites or with the inserted mutation into the predicted sites was used. Data shown are one representative of three independent experiments with similar results. (B) Promoter enzyme immunoassay using wild-type and mutated oligonucleotides within the FOXP3 promoter. Bars show mean ± SE of three independent experiments. (C) Chromatin immunoprecipitation assay results show binding of RUNX1 and RUNX3 complexes containing CBFβ to the human FOXP3 promoter in naive CD4+ T cells that were cultured with IL-2 together with anti-CD2/3/28 and TGF-β. There was no change in site occupancy in all immunoprecipitations when IGX1A negative control primers were used. The results are normalized to input and isotype control antibody. Bars show mean ± SE of three independent experiments. Statistical differences were verified by the paired Student's t test. *, P < 0.05
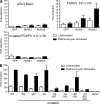
Figure 4.
Regulation of FOXP3 promoter activity by RUNX1 and RUNX3. (A) Human primary CD4+ cells were transfected with an empty vector (pGL3 Basic), a vector containing the wild-type or mutated FOXP3 promoter region (FOXP3 −511/+176) fused to the luciferase reporter gene together with a GFP, RUNX1, or RUNX3 expression vector. Bars show the mean luciferase activity ± SE measured as arbitrary light units of three independent experiments. (B) Human primary CD4+ cells were transfected with an empty vector (pGL3 Basic), a vector containing the putative FOXP3 promoter region (FOXP3 −511/+176) fused to the luciferase reporter gene, or with a vector containing the putative FOXP3 promoter region (FOXP3 −511/+176) with single RUNX binding sites mutated (53, 287, or 333) or with the combination of two or three RUNX binding sites mutated (53, 287, or 333) fused to the luciferase reporter gene. Bars show the mean luciferase activity ± SD measured as arbitrary light units of three independent experiments.
Figure 5.
Diminished capacity of _Cbfb_-deficient CD4-cre mice T cells in the generation of Foxp3+ CD4+ T cells. (A) FACS-purified naive CD4+ CD8− T cells from CbfbF/F CD4-cre and control CbfbF/+ CD4-cre mice were activated in vitro with anti-CD3/28 mAb, 50 U/ml IL-2, ± 10 nM retinoic acid (RA), and increasing concentrations of TGF-β. After 3 d in culture, the cells were restimulated with PMA + ionomycin, and then analyzed for intracellular Foxp3 and IFN-γ expression. One of five experiments is shown. (B) Naive CD4+ T cells from Cbfb CD4-cre or control mice (harboring a Foxp3-IRES-GFP allele) were adoptively transferred into Rag-deficient mice (5 × 106 cells per transfer). 6 wk later, TCRβ+CD4+ cells from the spleen, mesenteric lymph node (MLN), and lamina propria of the small intestine (LP) were analyzed for Foxp3-GFP expression. Results from one of four CbfbF/F CD4-cre and control CbfbF/+ CD4-cre mice with same findings are shown. The data from four sets of mice is shown in C. Statistical analysis was performed with Mann-Whitney U test. *, P < 0.05 between groups.
Figure 6.
CbfbF/F CD4-cre mouse cells and iT reg cells generated from human naive CD4+ T cells undergoing siRNA-mediated RUNX1 and RUNX3 knock down show a diminished suppressive activity. Experimental setup (A) and results of the mouse suppression assay (B), FACS-purified naive CD4+8− T cells from CbfbF/F CD4-cre (left) and control CbfbF/+ CD4-cre mice (Cd45.2; right) were activated in vitro with anti-CD3/28 mAb, 50 U/ml IL-2, and 2.5 ng/ml TGF-β. After 3 d, Foxp3-GFP+ cells were FACS-sorted and mixed with CFSE-loaded naive CD45.1+ CD4+ cells at the indicated ratios. These were then incubated with inactivated splenocytes and anti-CD3 mAb. After a further 4 d, CD45.1+ cells were analyzed for CFSE dilution. (C) As a control, CbfbF/+ CD4-cre CD4+ T cells activated in absence of TGF-β were mixed with CFSE-loaded naive CD45.1+ CD4+ cells at the indicated ratios. Four days later CD45.1+ cells were analyzed for CFSE dilution. The median division number (of the naive CD45.1+ CD4+ cells in the cultures containing Foxp3-GFP+ cells) is indicated in each of the histograms. One of three experiments is shown. (D) Human naive CD4+ T cells, transfected with RUNX1 and RUNX3 siRNA and cultured under iT reg differentiating conditions were used in an in vitro suppression assay, cultured together with autologous CFSE-labeled CD4+ T cells, and stimulated with anti-CD3 mAb. The CFSE dilution of the CD4+ T cell responder cells was analyzed after 5 d by flow cytometry. The T reg/responder CD4+ T cell ratios used were 1:20, 1:10, and 1:5. One of two experiments is shown.
Similar articles
- Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells.
Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S. Kitoh A, et al. Immunity. 2009 Oct 16;31(4):609-20. doi: 10.1016/j.immuni.2009.09.003. Epub 2009 Oct 1. Immunity. 2009. PMID: 19800266 - Runx1 and Runx3 are involved in the generation and function of highly suppressive IL-17-producing T regulatory cells.
Li L, Patsoukis N, Petkova V, Boussiotis VA. Li L, et al. PLoS One. 2012;7(9):e45115. doi: 10.1371/journal.pone.0045115. Epub 2012 Sep 12. PLoS One. 2012. PMID: 22984619 Free PMC article. - Oral tolerance induction with antigen conjugated to cholera toxin B subunit generates both Foxp3+CD25+ and Foxp3-CD25- CD4+ regulatory T cells.
Sun JB, Raghavan S, Sjöling A, Lundin S, Holmgren J. Sun JB, et al. J Immunol. 2006 Dec 1;177(11):7634-44. doi: 10.4049/jimmunol.177.11.7634. J Immunol. 2006. PMID: 17114433 - Interplay of transcription factors in T-cell differentiation and function: the role of Runx.
Wong WF, Kohu K, Chiba T, Sato T, Satake M. Wong WF, et al. Immunology. 2011 Feb;132(2):157-64. doi: 10.1111/j.1365-2567.2010.03381.x. Epub 2010 Nov 23. Immunology. 2011. PMID: 21091910 Free PMC article. Review. - Functional stability of Foxp3+ regulatory T cells.
da Silva Martins M, Piccirillo CA. da Silva Martins M, et al. Trends Mol Med. 2012 Aug;18(8):454-62. doi: 10.1016/j.molmed.2012.06.001. Epub 2012 Jul 6. Trends Mol Med. 2012. PMID: 22771168 Review.
Cited by
- The cnidarian origin of the proto-oncogenes NF-κB/STAT and WNT-like oncogenic pathway drives the ctenophores (Review).
Sinkovics JG. Sinkovics JG. Int J Oncol. 2015 Oct;47(4):1211-29. doi: 10.3892/ijo.2015.3102. Epub 2015 Jul 23. Int J Oncol. 2015. PMID: 26239915 Free PMC article. Review. - Increased DNA methylation variability in rheumatoid arthritis-discordant monozygotic twins.
Webster AP, Plant D, Ecker S, Zufferey F, Bell JT, Feber A, Paul DS, Beck S, Barton A, Williams FMK, Worthington J. Webster AP, et al. Genome Med. 2018 Sep 4;10(1):64. doi: 10.1186/s13073-018-0575-9. Genome Med. 2018. PMID: 30176915 Free PMC article. - Ultrastructural Localization and Molecular Associations of HCV Capsid Protein in Jurkat T Cells.
Fernández-Ponce C, Durán-Ruiz MC, Narbona-Sánchez I, Muñoz-Miranda JP, Arbulo-Echevarria MM, Serna-Sanz A, Baumann C, Litrán R, Aguado E, Bloch W, García-Cozar F. Fernández-Ponce C, et al. Front Microbiol. 2018 Jan 4;8:2595. doi: 10.3389/fmicb.2017.02595. eCollection 2017. Front Microbiol. 2018. PMID: 29354102 Free PMC article. - miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3.
Qiu YY, Zhang YW, Qian XF, Bian T. Qiu YY, et al. Am J Transl Res. 2017 Jul 15;9(7):3184-3199. eCollection 2017. Am J Transl Res. 2017. PMID: 28804539 Free PMC article. - Natural production and functional effects of alternatively spliced interleukin-4 protein in asthma.
Luzina IG, Lockatell V, Lavania S, Pickering EM, Kang PH, Bashkatova YN, Andreev SM, Atamas SP. Luzina IG, et al. Cytokine. 2012 Apr;58(1):20-6. doi: 10.1016/j.cyto.2011.12.017. Epub 2012 Jan 15. Cytokine. 2012. PMID: 22249152 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous