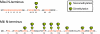Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi - PubMed (original) (raw)
Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Chen Chen et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Tudor domains are protein modules that mediate protein-protein interactions, potentially by binding to methylated ligands. A group of germline specific single and multiTudor domain containing proteins (TDRDs) represented by drosophila Tudor and its mammalian orthologs Tdrd1, Tdrd4/RNF17, and Tdrd6 play evolutionarily conserved roles in germinal granule/nuage formation and germ cell specification and differentiation. However, their physiological ligands, and the biochemical and structural basis for ligand recognition, are largely unclear. Here, by immunoprecipitation of endogenous murine Piwi proteins (Miwi and Mili) and proteomic analysis of complexes related to the piRNA pathway, we show that the TDRD group of Tudor proteins are physiological binding partners of Piwi family proteins. In addition, mass spectrometry indicates that arginine residues in RG repeats at the N-termini of Miwi and Mili are methylated in vivo. Notably, we found that Tdrkh/Tdrd2, a novel single Tudor domain containing protein identified in the Miwi complex, is expressed in the cytoplasm of male germ cells and directly associates with Miwi. Mutagenesis studies mapped the Miwi-Tdrkh interaction to the very N-terminal RG/RA repeats of Miwi and showed that the Tdrkh Tudor domain is critical for binding. Furthermore, we have solved the crystal structure of the Tdrkh Tudor domain, which revealed an aromatic binding pocket and negatively charged binding surface appropriate for accommodating methylated arginine. Our findings identify a methylation-directed protein interaction mechanism in germ cells mediated by germline Tudor domains and methylated Piwi family proteins, and suggest a complex mode of regulating the organization and function of Piwi proteins in piRNA silencing pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Proteomic analysis of endogenous Piwi complexes identifies Tudor domain family proteins as physiological binding partners. (A) Hierarchical clustering of proteins identified by 2 independent immunoprecipitations (IP) of Miwi, Mili, and corresponding IgG controls through tandem mass spectrometry. Proteins for which 3 or more peptides were identified are shown. Proteins that interact specifically with Miwi or Mili are illustrated in blue and red boxes, respectively. The cyan box indicates common proteins associated with both Miwi and Mili. (B) Identification of multiple germline Tudor domain proteins in Miwi and Mili complexes. Miwi and Mili protein interaction networks are shown with baits in blue and Tudor domain proteins in orange. Proteins represented by 3 or more peptides and not present in the IgG control IP, and proteins represented by peptides with a 5-fold increase over the IgG control IP are shown.
Fig. 2.
Arginine methylation sites detected on endogenous Miwi and Mili by mass spectrometry. N-terminal RG/RA-rich sequences are show in red. Identified methylation sites (Me) are shown above the relevant arginine, with the residue numbers underneath.
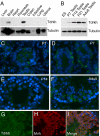
Fig. 3.
Expression kinetics and cellular localization of Tdrkh in the mouse testis. (A) Tissue distribution of Tdrkh protein by Western blotting. Tissue lysates were probed with anti-Tdrkh antibody; anti-Tubulin antibody was used as a control. (B) Western blot analysis of temporal Tdrkh protein expression in mouse testes. ES: mouse embryonic stem cell, P: postnatal day. (C–F) Immunofluorescence staining of Tdrkh (green) in testes of different developmental ages. P: postnatal day. (G–I) Colocalization of Tdrkh and Mvh in the adult testis.
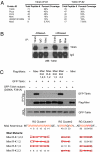
Fig. 4.
Tdrkh directly interacts with Miwi in vivo and in vitro through its Tudor domain. (A) Miwi is among the top specific interaction partners complexed with Tdrkh. Immunoprecipitation of Tdrkh from adult testis lysate and gel-free mass spectrometry were performed. Specific binding proteins are ranked based on the total peptide number identified. The top 5 Tdrkh interacting proteins with total peptide numbers and percentage of sequence coverage are shown for 2 independent immunoprecipitation experiments. (B) The interaction between Tdrkh and Miwi is RNA independent. Endogenous Tdrkh and Miwi were immunoprecipitated from adult testis lysates treated with or without RNaseA using anti-Tdrkh and anti-Miwi antibodies, respectively and immunoblotted with anti-Tdrkh antibody. (C) Tdrkh binds to the first cluster of RG/RA repeats on Miwi via its Tudor domain. HEK293T cells were cotransfected with Flag-Miwi or Flag-Miwi (R-K) mutants and GFP-Tdrkh or GFP-Tdrkh Tudor domain mutant (D390A, F391A). Flag-tagged protein complexes immunoprecipitated from cell extracts and whole cell lysates (WCL) were probed with anti-GFP and anti-Flag antibodies. The scheme of Miwi arginine mutations is shown in the bottom panel, with a red cross indicating an R-K mutant.
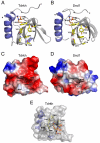
Fig. 5.
Crystal structure of the Tudor domain of Tdrkh. (A) Ribbon representation of the Tdrkh Tudor domain crystal structure. The residues comprising the aromatic binding pocket are shown in yellow. (B) Ribbon representation of the Snd1 Tudor domain crystal structure. (C) Surface representation of the Tdrkh Tudor domain crystal structure. (D) Surface representation of the Snd1 Tudor domain crystal structure. (E) Molecular docking of a GRG peptide with sDMA into the aromatic cage of the Tdrkh Tudor domain.
Similar articles
- Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Zhang H, Liu K, Izumi N, Huang H, Ding D, Ni Z, Sidhu SS, Chen C, Tomari Y, Min J. Zhang H, et al. Proc Natl Acad Sci U S A. 2017 Nov 21;114(47):12483-12488. doi: 10.1073/pnas.1711486114. Epub 2017 Nov 8. Proc Natl Acad Sci U S A. 2017. PMID: 29118143 Free PMC article. - Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members.
Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Vagin VV, et al. Genes Dev. 2009 Aug 1;23(15):1749-62. doi: 10.1101/gad.1814809. Epub 2009 Jul 7. Genes Dev. 2009. PMID: 19584108 Free PMC article. - MIWI N-terminal RG motif promotes efficient pachytene piRNA production and spermatogenesis independent of LINE1 transposon silencing.
Wei C, Jing J, Yan X, Mann JM, Geng R, Xie H, Demireva EY, Hess RA, Ding D, Chen C. Wei C, et al. PLoS Genet. 2023 Nov 13;19(11):e1011031. doi: 10.1371/journal.pgen.1011031. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 37956204 Free PMC article. - Structure and function of eTudor domain containing TDRD proteins.
Gan B, Chen S, Liu H, Min J, Liu K. Gan B, et al. Crit Rev Biochem Mol Biol. 2019 Apr;54(2):119-132. doi: 10.1080/10409238.2019.1603199. Epub 2019 May 3. Crit Rev Biochem Mol Biol. 2019. PMID: 31046474 Review. - Tudor domain-containing proteins of Drosophila melanogaster.
Ying M, Chen D. Ying M, et al. Dev Growth Differ. 2012 Jan;54(1):32-43. doi: 10.1111/j.1440-169x.2011.01308.x. Dev Growth Differ. 2012. PMID: 23741747 Review.
Cited by
- PNLDC1 catalysis and postnatal germline function are required for piRNA trimming, LINE1 silencing, and spermatogenesis in mice.
Wei C, Yan X, Mann JM, Geng R, Wang Q, Xie H, Demireva EY, Sun L, Ding D, Chen C. Wei C, et al. PLoS Genet. 2024 Sep 23;20(9):e1011429. doi: 10.1371/journal.pgen.1011429. eCollection 2024 Sep. PLoS Genet. 2024. PMID: 39312580 Free PMC article. - The functions and mechanisms of piRNAs in mediating mammalian spermatogenesis and their applications in reproductive medicine.
Du L, Chen W, Zhang D, Cui Y, He Z. Du L, et al. Cell Mol Life Sci. 2024 Sep 2;81(1):379. doi: 10.1007/s00018-024-05399-6. Cell Mol Life Sci. 2024. PMID: 39222270 Free PMC article. Review. - A Novel Compound Heterozygous Mutation in TDRD9 Causes Oligozoospermia.
Wang W, Feng Y, Dong J, Zhou Z, Jing J, Li Z, Chen L, Lin X, Ma J, Yao B. Wang W, et al. Reprod Sci. 2024 Nov;31(11):3413-3419. doi: 10.1007/s43032-024-01665-x. Epub 2024 Aug 22. Reprod Sci. 2024. PMID: 39174853 Free PMC article. - piRNA loading triggers MIWI translocation from the intermitochondrial cement to chromatoid body during mouse spermatogenesis.
Wei H, Gao J, Lin DH, Geng R, Liao J, Huang TY, Shang G, Jing J, Fan ZW, Pan D, Yin ZQ, Li T, Liu X, Zhao S, Chen C, Li J, Wang X, Ding D, Liu MF. Wei H, et al. Nat Commun. 2024 Mar 15;15(1):2343. doi: 10.1038/s41467-024-46664-3. Nat Commun. 2024. PMID: 38491008 Free PMC article. - PNLDC1 catalysis and postnatal germline function are required for piRNA trimming, LINE1 silencing, and spermatogenesis in mice.
Wei C, Yan X, Mann JM, Geng R, Xie H, Demireva EY, Sun L, Ding D, Chen C. Wei C, et al. bioRxiv [Preprint]. 2023 Dec 27:2023.12.26.573375. doi: 10.1101/2023.12.26.573375. bioRxiv. 2023. PMID: 38234819 Free PMC article. Updated. Preprint.
References
- Maurer-Stroh S, et al. The Tudor domain “royal family”: Tudor, plant agenet, chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. - PubMed
- Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochem Cell Biol. 2009;87:93–105. - PubMed
- Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. - PubMed
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases