Suppression of hypoxia-inducible factor 2alpha restores p53 activity via Hdm2 and reverses chemoresistance of renal carcinoma cells - PubMed (original) (raw)
Suppression of hypoxia-inducible factor 2alpha restores p53 activity via Hdm2 and reverses chemoresistance of renal carcinoma cells
Andrew M Roberts et al. Cancer Res. 2009.
Abstract
p53 mutations are rarely detected in clear cell renal cell carcinoma (CCRCC), but, paradoxically, these tumors remain highly resistant to chemotherapy and death receptor-induced death. Here, we show that the accumulation of hypoxia-inducible factor 2alpha (HIF2alpha), a critical oncogenic event in CCRCC following the loss of von Hippel-Lindau (VHL) tumor suppressor protein, leads to Hdm2-mediated suppression of p53. Primary CCRCC specimens exhibiting strong hypoxic signatures show increased levels of activated nuclear phospho-Hdm2(Ser(166)), which is concomitant with low p53 expression. The abrogation of Hdm2-p53 interaction using the small-molecule Hdm2 inhibitor nutlin-3 or the downregulation of HIF2alpha via HIF2alpha-specific short hairpin RNA or wild-type VHL reconstitution restores p53 function and reverses the resistance of CCRCC cells to Fas-mediated and chemotherapy-induced cell death. These findings unveil a mechanistic link between HIF2alpha and p53 and provide a rationale for combining Hdm2 antagonists with chemotherapy for the treatment of CCRCC.
Figures
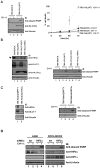
Figure 1. Activation of HIF2α is associated with resistance to Fas-mediated apoptosis
(A) 786-MOCK and 786-VHL(WT) cells treated with or without 50 ng/mL CH-11 were immunoblotted with the indicated antibodies (left panel) and cell viability measured by Trypan Blue exclusion assay (right panel). (B) Indicated 786-O subclones treated with (+) or without (−) 50 ng/mL CH-11 were immunoblotted with the indicated antibodies. * indicates non-specific protein band. (C) Total cell lysates prepared from 786-RetroshHIF2α or 786-RetroEMPTY treated with (+) or without (−) 50 ng/mL CH-11 were immunoblotted with the indicated antibodies. (D) A498 and RCC4-MOCK cells were transfected with scrambled siRNA (SCR) or HIF2α-siRNA (HIF2α), treated with (+) or without (−) 50 ng/mL CH-11 and equal amounts of total cell lysates immunoblotted with the indicated antibodies. IB: immunoblot.
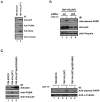
Figure 2. Resistance to Fas-mediated apoptosis is p53-dependent
(A) Equal amounts of total cell lysates from 786-MOCK and 786-VHL(WT) cells were immunoblotted with the indicated antibodies. (B) 786-VHL(WT) cells were transfected with scrambled (SCR) or p53-siRNA, treated with (+) or without (−) 50 ng/mL CH-11 and equal amounts of total cell lysates immunoblotted with the indicated antibodies. (C) Indicated 786-O subclones treated with (+) or without (−) 50 ng/mL CH-11 were immunoblotted with the indicated antibodies. IB: immunoblot.
Figure 3. HIF2α suppresses p53 via Akt-mediated activation of Hdm2
(A) Model of HIF2α-dependent regulation of p53 via Hdm2 (left panel). Equal amounts of total cell lysates from 786-MOCK and 786-VHL(WT) cells were immunoblotted with the indicated antibodies (right panel). (B) 786-MOCK cells treated with 5 μM nutlin-3 were immunoblotted with indicated antibodies. (C) 786-MOCK cells treated with 5 μM LY294002 were immunoblotted with indicated antibodies (left panel). 786-VHL(WT) cells were treated for 16 hours with conditioned media collected from 786-MOCK or 786-VHL(WT) maintained under normoxia or 786-VHL(WT) cells maintained under hypoxia. Total cell lysates were prepared and immunoblotted with the indicated antibodies (right panel). (D) Total cell lysates from 786-RetroEMPTY or 786-RetroshHIF2α cells were immunoblotted with the indicated antibodies. (A/C/D) Equal amounts of the same cell lysates were immunoprecipitated with anti-Hdm2 antibody and immunoblotted with anti-phospho-Hdm2(Ser166) antibody, which was subsequently stripped and re-immunoblotted with anti-Hdm2 antibody (bottom panels). IP: immunoprecipitation; IB: immunoblot.
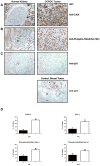
Figure 4. Primary CCRCC samples exhibit increased CAIX and phospho-Hdm2(Ser166) expression and low p53 expression
(A/B/C) Normal renal cortex (left panel) and CCRCC (right panel) were immunostained with the indicated antibodies. * indicates adjacent proximal convoluted tubule (A), proximal convoluted tubular epithelium (B) and tubular epithelium (C). Slim and block arrows represent scattered glomerular epithelial cells and mesangial cells, respectively (B). Invasive breast carcinoma served as a positive control for p53 staining (C) in which block and slim arrows represent adjacent adipocytes and non-tumor lobular breast tissue, respectively (bottom right panel). 400x original magnification. (D) Percentage of CCRCC tumors on TMA showing positive (+) or (−) p53 immunostaining (upper left panel). VHL status of p53-negative tumor samples (upper right panel). VHL (lower left panel) and p53 (lower right panel) status of phospho-Hdm2(Ser166)-positive tumor samples. Error bars indicate standard errors determined from two independent blind-scoring.
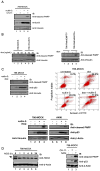
Figure 5. Suppression of HIF2α restores p53 activity and reverses chemoresistance of CCRCC
(A) 786-MOCK cells treated with (+) or without (−) 30 μM nutlin-3 or/and 50 ng/mL CH-11 were immunoblotted with the indicated antibodies. (B) 786-MOCK and 786-VHL(WT) cells treated with doxorubicin (dox) were immunoblotted with indicated antibodies (left panel). 786-RetroEMPTY and 786-RetroshHIF2α cells treated with (+) or without (−) 10 ng/mL dox were immunoblotted with indicated antibodies (right panel). (C) 786-MOCK cells treated with (+) or without (−) 30 μM nutlin-3 or/and 10 ng/mL dox were immunoblotted with indicated antibodies (top left panel) or stained with Annexin V-FITC and propidium iodide and analyzed by flow cytometry (top right panel). 786-MOCK and A498 cells treated with (+) or without (−) 30 μM nutlin-3 or/and 200 nM etoposide were immunoblotted with the indicated antibodies (bottom panel). (D) 786-MOCK cells treated with 640 ng/ml neocarzinostatin (NCS) were immunoblotted with the indicated antibodies. Arrow indicates Hdm2; * indicates non-specific protein band (left panel). 786-MOCK cells treated with (+) or without (−) 50 ng/mL CH-11 or 640 ng/ml NCS were immunoblotted with the indicated antibodies (right panel). IB: immunoblot.
Similar articles
- Low expression of phosphatase and tensin homolog in clear‑cell renal cell carcinoma contributes to chemoresistance through activating the Akt/HDM2 signaling pathway.
Chen J, Zhu H, Zhang Y, Cui MH, Han LY, Jia ZH, Wang L, Teng H, Miao LN. Chen J, et al. Mol Med Rep. 2015 Aug;12(2):2622-8. doi: 10.3892/mmr.2015.3740. Epub 2015 May 7. Mol Med Rep. 2015. PMID: 25954860 Free PMC article. - Chemotherapy-mediated p53-dependent DNA damage response in clear cell renal cell carcinoma: role of the mTORC1/2 and hypoxia-inducible factor pathways.
Selvarajah J, Nathawat K, Moumen A, Ashcroft M, Carroll VA. Selvarajah J, et al. Cell Death Dis. 2013 Oct 17;4(10):e865. doi: 10.1038/cddis.2013.395. Cell Death Dis. 2013. PMID: 24136229 Free PMC article. - Regulation of angiogenic factors by HDM2 in renal cell carcinoma.
Carroll VA, Ashcroft M. Carroll VA, et al. Cancer Res. 2008 Jan 15;68(2):545-52. doi: 10.1158/0008-5472.CAN-06-4738. Cancer Res. 2008. PMID: 18199551 - Targeting HIF-2 α in clear cell renal cell carcinoma: A promising therapeutic strategy.
Martínez-Sáez O, Gajate Borau P, Alonso-Gordoa T, Molina-Cerrillo J, Grande E. Martínez-Sáez O, et al. Crit Rev Oncol Hematol. 2017 Mar;111:117-123. doi: 10.1016/j.critrevonc.2017.01.013. Epub 2017 Jan 28. Crit Rev Oncol Hematol. 2017. PMID: 28259286 Review. - A Precision Strategy to Cure Renal Cell Carcinoma by Targeting Transglutaminase 2.
Kim SY, Keillor JW. Kim SY, et al. Int J Mol Sci. 2020 Apr 3;21(7):2493. doi: 10.3390/ijms21072493. Int J Mol Sci. 2020. PMID: 32260198 Free PMC article. Review.
Cited by
- Noncytotoxic differentiation treatment of renal cell cancer.
Negrotto S, Hu Z, Alcazar O, Ng KP, Triozzi P, Lindner D, Rini B, Saunthararajah Y. Negrotto S, et al. Cancer Res. 2011 Feb 15;71(4):1431-41. doi: 10.1158/0008-5472.CAN-10-2422. Epub 2011 Feb 8. Cancer Res. 2011. PMID: 21303982 Free PMC article. - Hypoxia-induced cytotoxic drug resistance in osteosarcoma is independent of HIF-1Alpha.
Adamski J, Price A, Dive C, Makin G. Adamski J, et al. PLoS One. 2013 Jun 13;8(6):e65304. doi: 10.1371/journal.pone.0065304. Print 2013. PLoS One. 2013. PMID: 23785417 Free PMC article. - Oroxylin A reverses hypoxia-induced cisplatin resistance through inhibiting HIF-1α mediated XPC transcription.
Liu Y, Wang X, Li W, Xu Y, Zhuo Y, Li M, He Y, Wang X, Guo Q, Zhao L, Qiang L. Liu Y, et al. Oncogene. 2020 Nov;39(45):6893-6905. doi: 10.1038/s41388-020-01474-x. Epub 2020 Sep 25. Oncogene. 2020. PMID: 32978517 - The effect of vorinostat on the development of resistance to doxorubicin in neuroblastoma.
Lautz TB, Jie C, Clark S, Naiditch JA, Jafari N, Qiu YY, Zheng X, Chu F, Madonna MB. Lautz TB, et al. PLoS One. 2012;7(7):e40816. doi: 10.1371/journal.pone.0040816. Epub 2012 Jul 19. PLoS One. 2012. PMID: 22829886 Free PMC article. - RING finger protein 31 promotes p53 degradation in breast cancer cells.
Zhu J, Zhao C, Zhuang T, Jonsson P, Sinha I, Williams C, Strömblad S, Dahlman-Wright K. Zhu J, et al. Oncogene. 2016 Apr 14;35(15):1955-64. doi: 10.1038/onc.2015.260. Epub 2015 Jul 6. Oncogene. 2016. PMID: 26148235 Free PMC article.
References
- Cohen HT, McGovern FJ. Renal-cell carcinoma. The New England journal of medicine. 2005;353(23):2477–90. - PubMed
- Brugarolas J. Renal-cell carcinoma--molecular pathways and therapies. The New England journal of medicine. 2007;356(2):185–7. - PubMed
- Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108(2):153–64. - PubMed
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes & development. 1996;10(9):1054–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CAPMC/ CIHR/Canada
- R01 CA046677-16A2/CA/NCI NIH HHS/United States
- R01 CA046677-15S1/CA/NCI NIH HHS/United States
- R01 CA046677-15/CA/NCI NIH HHS/United States
- CA046677/CA/NCI NIH HHS/United States
- R01 CA046677/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous