Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov.and Propionivibrio militaris, sp. nov - PubMed (original) (raw)
Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov.and Propionivibrio militaris, sp. nov
J Cameron Thrash et al. Appl Microbiol Biotechnol. 2010 Mar.
Abstract
Novel dissimilatory perchlorate-reducing bacteria (DPRB) were isolated from enrichments conducted under conditions different from those of all previously described DPRB. Strain LT-1(T) was enriched using medium buffered at pH 6.6 with 2-(N-morpholino)ethanesulfonic acid (MES) and had only 95% 16S rRNA gene identity with its closest relative, Azonexus caeni. Strain MP(T) was enriched in the cathodic chamber of a perchlorate-reducing bioelectrical reactor (BER) and together with an additional strain, CR (99% 16S rRNA gene identity), had 97% 16S rRNA gene identity with Propionivibrio limicola. The use of perchlorate and other electron acceptors distinguished strains MP(T) and CR from P. limicola physiologically. Strain LT-1(T) had differences in electron donor utilization and optimum growth temperatures from A. caeni. Strains LT-1(T) and MP(T) are the first DPRB to be described in the Betaproteobacteria outside of the Dechloromonas and Azospira genera. On the basis of phylogenetic and physiological features, strain LT-1(T) represents a novel genus in the Rhodocyclaceae; strain MP(T) represents a novel species within the genus Propionivibrio. The names Dechlorobacter hydrogenophilus gen. nov., sp. nov and Propionivibrio militaris sp. nov. are proposed.
Figures
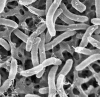
Fig. 1
SEM of strain MPT, scale bar = 1 µm
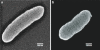
Fig. 2
SEMs of strain LT-1T, scale bars = 125 nm
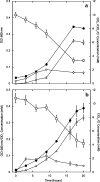
Fig. 3
Growth curves of strain MPT (a) and strain LT-1T (b). Cell number increase was monitored with optical density (OD) at 600 nm. Closed circles OD, open squares [ClO4−], open triangles [Cl−], open diamonds, [ClO3−]; all concentrations in millimolar. Error bars represent the standard deviation of triplicate experiments
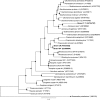
Fig. 4
Bayesian 16S rRNA gene phylogenetic tree showing the position of strains MPT, CR, and LT-1T in the Rhodocylaceae. Scale bar represents 0.07 changes per position. Circles at the nodes indicate posterior probabilities >0.85 (closed) or <0.85 (open)
Similar articles
- Magnetospirillum bellicus sp. nov., a novel dissimilatory perchlorate-reducing alphaproteobacterium isolated from a bioelectrical reactor.
Thrash JC, Ahmadi S, Torok T, Coates JD. Thrash JC, et al. Appl Environ Microbiol. 2010 Jul;76(14):4730-7. doi: 10.1128/AEM.00015-10. Epub 2010 May 21. Appl Environ Microbiol. 2010. PMID: 20495050 Free PMC article. - Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position.
Achenbach LA, Michaelidou U, Bruce RA, Fryman J, Coates JD. Achenbach LA, et al. Int J Syst Evol Microbiol. 2001 Mar;51(Pt 2):527-533. doi: 10.1099/00207713-51-2-527. Int J Syst Evol Microbiol. 2001. PMID: 11321099 - Description of Pelomonas aquatica sp. nov. and Pelomonas puraquae sp. nov., isolated from industrial and haemodialysis water.
Gomila M, Bowien B, Falsen E, Moore ERB, Lalucat J. Gomila M, et al. Int J Syst Evol Microbiol. 2007 Nov;57(Pt 11):2629-2635. doi: 10.1099/ijs.0.65149-0. Int J Syst Evol Microbiol. 2007. PMID: 17978231 - Dechloromonas hortensis sp. nov. and strain ASK-1, two novel (per)chlorate-reducing bacteria, and taxonomic description of strain GR-1.
Wolterink A, Kim S, Muusse M, Kim IS, Roholl PJM, van Ginkel CG, Stams AJM, Kengen SWM. Wolterink A, et al. Int J Syst Evol Microbiol. 2005 Sep;55(Pt 5):2063-2068. doi: 10.1099/ijs.0.63404-0. Int J Syst Evol Microbiol. 2005. PMID: 16166710 - Tetrathiobacter kashmirensis gen. nov., sp. nov., a novel mesophilic, neutrophilic, tetrathionate-oxidizing, facultatively chemolithotrophic betaproteobacterium isolated from soil from a temperate orchard in Jammu and Kashmir, India.
Ghosh W, Bagchi A, Mandal S, Dam B, Roy P. Ghosh W, et al. Int J Syst Evol Microbiol. 2005 Sep;55(Pt 5):1779-1787. doi: 10.1099/ijs.0.63595-0. Int J Syst Evol Microbiol. 2005. PMID: 16166666
Cited by
- Identification of a parasitic symbiosis between respiratory metabolisms in the biogeochemical chlorine cycle.
Barnum TP, Cheng Y, Hill KA, Lucas LN, Carlson HK, Coates JD. Barnum TP, et al. ISME J. 2020 May;14(5):1194-1206. doi: 10.1038/s41396-020-0599-1. Epub 2020 Feb 5. ISME J. 2020. PMID: 32024948 Free PMC article. - Bioelectricity generation using long-term operated biocathode: RFLP based microbial diversity analysis.
Ramanaiah SV, Cordas CM, Matias SC, Reddy MV, Leitão JH, Fonseca LP. Ramanaiah SV, et al. Biotechnol Rep (Amst). 2021 Dec 5;32:e00693. doi: 10.1016/j.btre.2021.e00693. eCollection 2021 Dec. Biotechnol Rep (Amst). 2021. PMID: 34917493 Free PMC article. - (Per)chlorate reduction by an acetogenic bacterium, Sporomusa sp., isolated from an underground gas storage.
Balk M, Mehboob F, van Gelder AH, Rijpstra WI, Damsté JS, Stams AJ. Balk M, et al. Appl Microbiol Biotechnol. 2010 Sep;88(2):595-603. doi: 10.1007/s00253-010-2788-8. Epub 2010 Aug 3. Appl Microbiol Biotechnol. 2010. PMID: 20680263 Free PMC article. - The Perchlorate Reduction Genomic Island: Mechanisms and Pathways of Evolution by Horizontal Gene Transfer.
Melnyk RA, Coates JD. Melnyk RA, et al. BMC Genomics. 2015 Oct 26;16:862. doi: 10.1186/s12864-015-2011-5. BMC Genomics. 2015. PMID: 26502901 Free PMC article. - Fermentative Bacteria Influence the Competition between Denitrifiers and DNRA Bacteria.
van den Berg EM, Elisário MP, Kuenen JG, Kleerebezem R, van Loosdrecht MCM. van den Berg EM, et al. Front Microbiol. 2017 Sep 5;8:1684. doi: 10.3389/fmicb.2017.01684. eCollection 2017. Front Microbiol. 2017. PMID: 28928722 Free PMC article.
References
- Achenbach LA, Bender KS, Sun Y, Coates JD. The biochemistry and genetics of microbial perchlorate reduction. In: Gu B, Coates JD, editors. Perchlorate: environmental occurrence, interactions and treatment. New York: Springer; 2006. pp. 297–310.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous