Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro - PubMed (original) (raw)
Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro
Yiu-Wing Kam et al. PLoS One. 2009.
Abstract
Background: Entry of enveloped viruses into host cells requires the activation of viral envelope glycoproteins through cleavage by either intracellular or extracellular proteases. In order to gain insight into the molecular basis of protease cleavage and its impact on the efficiency of viral entry, we investigated the susceptibility of a recombinant native full-length S-protein trimer (triSpike) of the severe acute respiratory syndrome coronavirus (SARS-CoV) to cleavage by various airway proteases.
Methodology/principal findings: PURIFIED TRISPIKE PROTEINS WERE READILY CLEAVED IN VITRO BY THREE DIFFERENT AIRWAY PROTEASES: trypsin, plasmin and TMPRSS11a. High Performance Liquid Chromatography (HPLC) and amino acid sequencing analyses identified two arginine residues (R667 and R797) as potential protease cleavage site(s). The effect of protease-dependent enhancement of SARS-CoV infection was demonstrated with ACE2 expressing human bronchial epithelial cells 16HBE. Airway proteases regulate the infectivity of SARS-CoV in a fashion dependent on previous receptor binding. The role of arginine residues was further shown with mutant constructs (R667A, R797A or R797AR667A). Mutation of R667 or R797 did not affect the expression of S-protein but resulted in a differential efficacy of pseudotyping into SARS-CoVpp. The R667A SARS-CoVpp mutant exhibited a lack of virus entry enhancement following protease treatment.
Conclusions/significance: These results suggest that SARS S-protein is susceptible to airway protease cleavage and, furthermore, that protease mediated enhancement of virus entry depends on specific conformation of SARS S-protein upon ACE2 binding. These data have direct implications for the cell entry mechanism of SARS-CoV along the respiratory system and, furthermore expand the possibility of identifying potential therapeutic agents against SARS-CoV.
Conflict of interest statement
Competing Interests: Dr Lisa F. P. NG is a PLoS ONE Academic editor
Figures
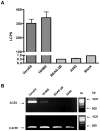
Figure 1. Susceptibility of various human airway epithelial cells to SARS-CoV S-mediated infection.
A, VeroE6 (10,000 cells/well), 16HBE, BEAS-2B and A549 (20,000 cells/well) cells were seeded onto 96-well plates 24 h before SARS-CoVpp infection. Pseudotypes (SARS-CoVpp) were collected from culture medium and concentrated as described previously . SARS-CoVpp were incubated with various cell lines and transduction was measured by determination of the luciferase activity expressed as luminescence counts per second (LCPS). VeroE6 cells were used as positive control. All experiments were performed in triplicates and data are presented as means±SE of two or three independent experiments. B, ACE2 expression from various mammalian airway cell lines. Cell lysates were collected and ACE2 RNA molecules were detected by RT-PCR. Amplified ACE2 cDNA products from 16HBE, BEAS-2B and A549 are shown in lanes 2 to 4, respectively. VeroE6 cell line (lane 1) was used as positive control for ACE2 expression. The quantity of total RNA templates was normalized to β-actin expression as shown from the lower panel. Lane M represents the DNA size marker and the size of DNA bands are indicated on the right.
Figure 2. Identification of airway protease cleavage site(s) along the amino acid sequence of SARS-CoV S glycoprotein.
A, Purified triSpike proteins (lane 1: detected by Western immunoblot, lane 2: silver staining) were incubated with 0.2 mU of trypsin (lane 3), plasmin (lane 4) or TMPRSS11a (lane 5) identified and purified from lungs and bronchi. Cleavage products were visualized and prepared as described in Materials and Methods. Amino acid sequences (T1, T2, P1, P2, N1, and N2) corresponding to the cleaved triSpike proteins are shown in the lower panel. B, Three different types of airway proteases (trypsin, plasmin and TMPRSS11a) utilize the same amino acid residues for protein cleavage. A schematic diagram representing the amino acid sequence of SARS-CoV S glycoprotein shows on top of the figure. A red circle indicates the location of potential cleavage site along the Spike glycoprotein. Red dots represent the basic amino acids, potential protease cleavage sites (red letter) and two red arrows indicate the cleavage sites identified. NTD – N-terminal domain, RBD – receptor-binding domain, RBM – receptor-binding motif, FP – fusion peptide, HR-N – N-terminal of heptad-repeat, HR-C – C-terminal of heptad-repeat, IC – Intracellular tail.
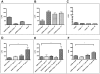
Figure 3. Effect of airway proteases treatment on SARS-CoVpp infectivity.
A, SARS-CoVpp was pre-incubated with either trypsin (T) or plasmin (P) (10 µg/ml) at 37°C for 20 min. Luciferase activity (LCPS) was measured from infected 16HBE cells. Asterisk (*) indicates a value of p<0.05 in two-tailed t tests. Experiments were performed in triplicates and values were expressed as means±SE from two independent experiments. SARS-CoVpp entry into susceptible cell lines was enhanced with the presence of airway proteases. B & C, Equal amounts of SARS-CoVpp and empp (normalized to p24 quantity) were pre-incubated with 16HBE cells on ice for 30 min. Cells were washed twice to remove any unbound pp. Cells were incubated with 10 µg/ml of either trypsin (T), TMPRSS11a (N) or 100 µg/ml of plasmin (P) at room temperature for 40 min. Luciferase activity (LCPS) was measured from infected 16HBE cells. Experiments were performed in triplicates and values were expressed as means±SE from two independent experiments. D-F, Next, SARS-CoVpp was pre-incubated with 16HBE cells on ice for 30 min. Cells were washed twice to remove any unbound pp. Cells were incubated with various concentrations of trypsin (T), plasmin (P) or TMPRSS11a (N) at room temperature for 40 min. Luciferase activity (LCPS) was measured from infected 16HBE cells. Asterisk (**) indicates a value of p<0.01 in two-tailed t tests. Experiments were performed in triplicates and values were expressed as means±SE from three independent experiments.
Figure 4. Differential expression of wild-type and mutant SARS S glycoprotein and SARS-CoV pseudotype production from 293T cells.
A, Different putative spike glycoprotein sequences from SARS-CoV isolates were obtained from NCBI. Name of SARS-CoV isolates and GenBank accession numbers are listed. Potential airway protease cleavage residues are highlighted in green. B, Three mutant constructs were made from the wild-type SARS-CoV Spike glycoprotein cDNA cloned in the vector pcDNA3.1 as described in Materials and Methods. C, Cell lysates were collected from 293T cells 48 h post transfection. Pseudotypes were collected from culture medium and concentrated as described previously . Samples (cell lysates or pseduotypes) were denatured, reduced and analyzed by 4–12% Bis-Tris SDS-PAGE gel and Western Blot using M2 monoclonal antibody against the FLAG peptide. Sizes of molecular weight markers are indicated on the right. Wild-type SARS spike (lane 1), R667A (lane 2), R797A (lane 3), R797AR667A (lane 4) and pseudotype without envelope (lane 5). D, Analysis of various types of SARS-CoVpp for viral entry. SARS-CoVpp (wild-type or mutant SARS S glycoprotein) were incubated with 16HBE cells and transduction was measured by determination of the luciferase activity (LCPS). Experiments were performed in duplicates and data are presented as means±SE from two independent experiments.
Figure 5. Role of amino acid residue 667 enhances SARS-CoVpp entry in the presence of airway proteases.
SARS-CoVpp (A) or R667App (B) was pre-incubated with 16HBE cells on ice for 30 min. Cells were washed twice to remove any unbound pp. Cells were incubated with 10 µg/ml of either trypsin (T), TMPRSS11a (N) or 100 µg/ml of plasmin (P) at room temperature for 40 min. Luciferase activity (LCPS) was measured from infected 16HBE cells. Asterisk (*) indicates a value of p<0.05 and (**) indicates a value of p<0.01 in two-tailed t tests. Experiments were performed in triplicates and values were expressed as means±SE from three independent experiments.
Similar articles
- Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease.
Bertram S, Glowacka I, Müller MA, Lavender H, Gnirss K, Nehlmeier I, Niemeyer D, He Y, Simmons G, Drosten C, Soilleux EJ, Jahn O, Steffen I, Pöhlmann S. Bertram S, et al. J Virol. 2011 Dec;85(24):13363-72. doi: 10.1128/JVI.05300-11. Epub 2011 Oct 12. J Virol. 2011. PMID: 21994442 Free PMC article. - Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63.
Glowacka I, Bertram S, Herzog P, Pfefferle S, Steffen I, Muench MO, Simmons G, Hofmann H, Kuri T, Weber F, Eichler J, Drosten C, Pöhlmann S. Glowacka I, et al. J Virol. 2010 Jan;84(2):1198-205. doi: 10.1128/JVI.01248-09. Epub 2009 Oct 28. J Virol. 2010. PMID: 19864379 Free PMC article. - Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2.
Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Matsuyama S, et al. J Virol. 2010 Dec;84(24):12658-64. doi: 10.1128/JVI.01542-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926566 Free PMC article. - SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium.
Sims AC, Burkett SE, Yount B, Pickles RJ. Sims AC, et al. Virus Res. 2008 Apr;133(1):33-44. doi: 10.1016/j.virusres.2007.03.013. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451829 Free PMC article. Review. - Insights from the association of SARS-CoV S-protein with its receptor, ACE2.
Li W, Choe H, Farzan M. Li W, et al. Adv Exp Med Biol. 2006;581:209-18. doi: 10.1007/978-0-387-33012-9_36. Adv Exp Med Biol. 2006. PMID: 17037532 Free PMC article. Review. No abstract available.
Cited by
- Furin: A Potential Therapeutic Target for COVID-19.
Wu C, Zheng M, Yang Y, Gu X, Yang K, Li M, Liu Y, Zhang Q, Zhang P, Wang Y, Wang Q, Xu Y, Zhou Y, Zhang Y, Chen L, Li H. Wu C, et al. iScience. 2020 Oct 23;23(10):101642. doi: 10.1016/j.isci.2020.101642. Epub 2020 Oct 5. iScience. 2020. PMID: 33043282 Free PMC article. - A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors.
Singh M, Bansal V, Feschotte C. Singh M, et al. SSRN [Preprint]. 2020 May 27:3611279. doi: 10.2139/ssrn.3611279. SSRN. 2020. PMID: 32714119 Free PMC article. Updated. Preprint. - Competitive cleavage of SARS-CoV-2 spike protein and epithelial sodium channel by plasmin as a potential mechanism for COVID-19 infection.
Hou Y, Yu T, Wang T, Ding Y, Cui Y, Nie H. Hou Y, et al. Am J Physiol Lung Cell Mol Physiol. 2022 Nov 1;323(5):L569-L577. doi: 10.1152/ajplung.00152.2022. Epub 2022 Oct 4. Am J Physiol Lung Cell Mol Physiol. 2022. PMID: 36193902 Free PMC article. - Neutrophil Extracellular Traps Induce the Epithelial-Mesenchymal Transition: Implications in Post-COVID-19 Fibrosis.
Pandolfi L, Bozzini S, Frangipane V, Percivalle E, De Luigi A, Violatto MB, Lopez G, Gabanti E, Carsana L, D'Amato M, Morosini M, De Amici M, Nebuloni M, Fossali T, Colombo R, Saracino L, Codullo V, Gnecchi M, Bigini P, Baldanti F, Lilleri D, Meloni F. Pandolfi L, et al. Front Immunol. 2021 Jun 14;12:663303. doi: 10.3389/fimmu.2021.663303. eCollection 2021. Front Immunol. 2021. PMID: 34194429 Free PMC article. - A Revisit to the Research Updates of Drugs, Vaccines, and Bioinformatics Approaches in Combating COVID-19 Pandemic.
Sumon TA, Hussain MA, Hasan MT, Hasan M, Jang WJ, Bhuiya EH, Chowdhury AAM, Sharifuzzaman SM, Brown CL, Kwon HJ, Lee EW. Sumon TA, et al. Front Mol Biosci. 2021 Jan 25;7:585899. doi: 10.3389/fmolb.2020.585899. eCollection 2020. Front Mol Biosci. 2021. PMID: 33569389 Free PMC article. Review.
References
- Klenk HD, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. - PubMed
- McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, et al. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. - PubMed
- Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–454. - PubMed
- Kido H, Murakami M, Oba K, Chen Y, Towatari T. Cellular proteinases trigger the infectivity of the influenza A and Sendai viruses. Mol Cells. 1999;9:235–244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous