Profiling critical cancer gene mutations in clinical tumor samples - PubMed (original) (raw)
. 2009 Nov 18;4(11):e7887.
doi: 10.1371/journal.pone.0007887.
Catarina D Campbell, Sarah M Kehoe, Adam J Bass, Charles Hatton, Lili Niu, Matt Davis, Keluo Yao, Megan Hanna, Chandrani Mondal, Lauren Luongo, Caroline M Emery, Alissa C Baker, Juliet Philips, Deborah J Goff, Michelangelo Fiorentino, Mark A Rubin, Kornelia Polyak, Jennifer Chan, Yuexiang Wang, Jonathan A Fletcher, Sandro Santagata, Gianni Corso, Franco Roviello, Ramesh Shivdasani, Mark W Kieran, Keith L Ligon, Charles D Stiles, William C Hahn, Matthew L Meyerson, Levi A Garraway
Affiliations
- PMID: 19924296
- PMCID: PMC2774511
- DOI: 10.1371/journal.pone.0007887
Profiling critical cancer gene mutations in clinical tumor samples
Laura E MacConaill et al. PLoS One. 2009.
Erratum in
- PLoS One. 2010;5(9). doi: 10.1371/annotation/3a0c8fee-57ef-43ed-b6c2-55b503e6db5e
- PLoS One. 2010;5(9). doi: 10.1371/annotation/613c7509-e4c9-42ac-82fb-fc504400d9e0
Abstract
Background: Detection of critical cancer gene mutations in clinical tumor specimens may predict patient outcomes and inform treatment options; however, high-throughput mutation profiling remains underdeveloped as a diagnostic approach. We report the implementation of a genotyping and validation algorithm that enables robust tumor mutation profiling in the clinical setting.
Methodology: We developed and implemented an optimized mutation profiling platform ("OncoMap") to interrogate approximately 400 mutations in 33 known oncogenes and tumor suppressors, many of which are known to predict response or resistance to targeted therapies. The performance of OncoMap was analyzed using DNA derived from both frozen and FFPE clinical material in a diverse set of cancer types. A subsequent in-depth analysis was conducted on histologically and clinically annotated pediatric gliomas. The sensitivity and specificity of OncoMap were 93.8% and 100% in fresh frozen tissue; and 89.3% and 99.4% in FFPE-derived DNA. We detected known mutations at the expected frequencies in common cancers, as well as novel mutations in adult and pediatric cancers that are likely to predict heightened response or resistance to existing or developmental cancer therapies. OncoMap profiles also support a new molecular stratification of pediatric low-grade gliomas based on BRAF mutations that may have immediate clinical impact.
Conclusions: Our results demonstrate the clinical feasibility of high-throughput mutation profiling to query a large panel of "actionable" cancer gene mutations. In the future, this type of approach may be incorporated into both cancer epidemiologic studies and clinical decision making to specify the use of many targeted anticancer agents.
Conflict of interest statement
Competing Interests: In terms of financial disclosures, Levi Garraway is a consultant for and/or receives sponsored research from Novartis, Inc. None of these relationships constitute a conflict of interest for this work. This does not alter his adherence to all PLoS ONE policies on data sharing and materials.
Figures
Figure 1. The OncoMap process and performance in fresh frozen and FFPE-derived DNA.
A. An overview of the OncoMap process from tumor to mutation profile. See text for details. B. Receiver operator characteristic curves (ROCs) show the sensitivity and specificity for various cutoff values on the sample score of the validation samples. ROCs are plotted for fresh frozen (left) and FFPE-derived (right) DNAs, using bidirectional KRAS assays and Illumina data as a truth-set (see Methods S1).
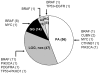
Figure 2. BRAFV600E mutations detected in archival samples of pediatric gliomas.
Abbreviations: PA - pilocytic astrocytoma, WHO grade I; LGG, nos – low-grade glioma, not otherwise specified, WHO grade I or II; GG – ganglioglioma; A2 – astrocytoma, WHO grade II; HG – high-grade glioma, WHO grade III or IV. Parentheses indicate total number of samples (in chart) or number of samples with indicated mutation (outside chart).
Similar articles
- High throughput interrogation of somatic mutations in high grade serous cancer of the ovary.
Matulonis UA, Hirsch M, Palescandolo E, Kim E, Liu J, van Hummelen P, MacConaill L, Drapkin R, Hahn WC. Matulonis UA, et al. PLoS One. 2011;6(9):e24433. doi: 10.1371/journal.pone.0024433. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931712 Free PMC article. - Ultrasensitive detection and identification of BRAF V600 mutations in fresh frozen, FFPE, and plasma samples of melanoma patients by E-ice-COLD-PCR.
How-Kit A, Lebbé C, Bousard A, Daunay A, Mazaleyrat N, Daviaud C, Mourah S, Tost J. How-Kit A, et al. Anal Bioanal Chem. 2014 Sep;406(22):5513-20. doi: 10.1007/s00216-014-7975-5. Epub 2014 Jun 27. Anal Bioanal Chem. 2014. PMID: 24969466 - High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed PCR and next-generation sequencing.
Bourgon R, Lu S, Yan Y, Lackner MR, Wang W, Weigman V, Wang D, Guan Y, Ryner L, Koeppen H, Patel R, Hampton GM, Amler LC, Wang Y. Bourgon R, et al. Clin Cancer Res. 2014 Apr 15;20(8):2080-91. doi: 10.1158/1078-0432.CCR-13-3114. Epub 2014 Feb 26. Clin Cancer Res. 2014. PMID: 24573554 - A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes.
Fumagalli D, Gavin PG, Taniyama Y, Kim SI, Choi HJ, Paik S, Pogue-Geile KL. Fumagalli D, et al. BMC Cancer. 2010 Mar 16;10:101. doi: 10.1186/1471-2407-10-101. BMC Cancer. 2010. PMID: 20233444 Free PMC article. - Use of a High-Throughput Genotyping Platform (OncoMap) for RAS Mutational Analysis to Predict Cetuximab Efficacy in Patients with Metastatic Colorectal Cancer.
Kim D, Hong YS, Kim JE, Kim KP, Lee JL, Chun SM, Kim J, Jang SJ, Kim TW. Kim D, et al. Cancer Res Treat. 2017 Jan;49(1):37-43. doi: 10.4143/crt.2016.069. Epub 2016 Apr 27. Cancer Res Treat. 2017. PMID: 27121720 Free PMC article.
Cited by
- Review of low-grade gliomas in children--evolving molecular era and therapeutic insights.
Khatua S, Wang J, Rajaram V. Khatua S, et al. Childs Nerv Syst. 2015 May;31(5):643-52. doi: 10.1007/s00381-015-2653-2. Epub 2015 Feb 27. Childs Nerv Syst. 2015. PMID: 25722047 Review. - Clinical bioinformatics desiderata for molecular tumor boards.
Pallocca M, Betti M, Baldinelli S, Palombo R, Bucci G, Mazzarella L, Tonon G, Ciliberto G. Pallocca M, et al. Brief Bioinform. 2024 Jul 25;25(5):bbae447. doi: 10.1093/bib/bbae447. Brief Bioinform. 2024. PMID: 39297878 Free PMC article. Review. - Molecular typing of Meningiomas by Desorption Electrospray Ionization Mass Spectrometry Imaging for Surgical Decision-Making.
Calligaris D, Feldman DR, Norton I, Brastianos PK, Dunn IF, Santagata S, Agar NY. Calligaris D, et al. Int J Mass Spectrom. 2015 Feb 1;377:690-698. doi: 10.1016/j.ijms.2014.06.024. Int J Mass Spectrom. 2015. PMID: 25844057 Free PMC article. - Clinical analysis and interpretation of cancer genome data.
Van Allen EM, Wagle N, Levy MA. Van Allen EM, et al. J Clin Oncol. 2013 May 20;31(15):1825-33. doi: 10.1200/JCO.2013.48.7215. Epub 2013 Apr 15. J Clin Oncol. 2013. PMID: 23589549 Free PMC article. Review. - Molecular testing in melanoma.
Wilson MA, Nathanson KL. Wilson MA, et al. Cancer J. 2012 Mar-Apr;18(2):117-23. doi: 10.1097/PPO.0b013e31824f11bf. Cancer J. 2012. PMID: 22453011 Free PMC article. Review.
References
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
- Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. - PubMed
- Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622–2629. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials