Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies - PubMed (original) (raw)
Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies
Brian L Browning et al. Am J Hum Genet. 2009 Dec.
Abstract
We present a novel method for simultaneous genotype calling and haplotype-phase inference. Our method employs the computationally efficient BEAGLE haplotype-frequency model, which can be applied to large-scale studies with millions of markers and thousands of samples. We compare genotype calls made with our method to genotype calls made with the BIRDSEED, CHIAMO, GenCall, and ILLUMINUS genotype-calling methods, using genotype data from the Illumina 550K and Affymetrix 500K arrays. We show that our method has higher genotype-call accuracy and yields fewer uncalled genotypes than competing methods. We perform single-marker analysis of data from the Wellcome Trust Case Control Consortium bipolar disorder and type 2 diabetes studies. For bipolar disorder, the genotype calls in the original study yield 25 markers with apparent false-positive association with bipolar disorder at a p < 10(-7) significance level, whereas genotype calls made with our method yield no associated markers at this significance threshold. Conversely, for markers with replicated association with type 2 diabetes, there is good concordance between genotype calls used in the original study and calls made by our method. Results from single-marker and haplotypic analysis of our method's genotype calls for the bipolar disorder study indicate that our method is highly effective at eliminating genotyping artifacts that cause false-positive associations in genome-wide association studies. Our new genotype-calling methods are implemented in the BEAGLE and BEAGLECALL software packages.
Figures
Figure 1
A Schematic of the Proposed Method for Simultaneous Genotype Calling and Haplotype-Phase Inference
Figure 2
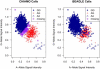
Allele Signal Intensities and Genotype Calls for Marker rs4242382 Affymetrix 500K chip allele signal intensities, CHIAMO genotype calls (left panel), and BEAGLE genotype calls (right panel) for marker rs4242382 for 1373 individuals from the 58BC cohort that were genotyped on the Affymetrix 500K chip and the Illumina 550K chip and passed genome-wide QC filters (see Material and Methods). Genotypes with CHIAMO posterior probability < 0.90 and BEAGLE posterior probability < 0.97 are labeled as missing. Genotype calls for these samples made with the use of Illumina 550K chip data have 96.2% concordance with CHIAMO genotype calls and 99.9% concordance with BEAGLE genotype calls.
Figure 3
Genotype Discordance and Missing-Data Rates Discordance rates for genotype calls for autosomal Affymetrix 500K chip data (left panel) and autosomal Illumina 550K chip data (right panel) are computed with the use of high-confidence genotype calls (probability > 0.999995) from the alternate platform. The genotype discordance rate and missing-data rate depend on the quality-score threshold required for calling a genotype. For each method and each possible calling threshold, the proportion of missing genotypes and the discordance rate for called genotypes was computed. The discordance and missing-data rates corresponding to calling thresholds of 0.9, 0.99, and 0.999 posterior genotype probability are shown for the genotype-calling methods that report genotype probabilities.
Figure 4
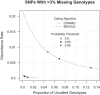
Genotype Discordance and Missing-Data Rates at SNPs with > 3% Missing CHIAMO Genotypes Discordance and missing-data rates are given for CHIAMO and BEAGLE Affymetrix 500K chip genotype calls for the subset of SNPs with > 3% missing CHIAMO genotypes. Discordance rates are computed with the use of high-confidence (genotype probability > 0.999995) BEAGLE Illumina 550K chip genotype calls. The unfilled triangle, filled square, and filled triangle identify the discordance and missing-data rates corresponding to calling thresholds of 0.9, 0.95, and 0.99 posterior genotype probability.
Figure 5
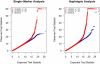
Quantile-Quantile Plots for Single-Marker and Haplotypic Analyses of Bipolar Disorder Expected and observed association chi-square test statistics from analysis of CHIAMO genotype calls and BEAGLE genotype calls of WTCCC bipolar disorder and control data. An allelic test statistic and three genotypic test statistics, corresponding to dominant, overdominant, and recessive models, are computed for each marker (left panel) and each tested haplotype cluster (right panel).
Figure 6
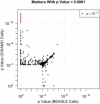
p Values from Single-Marker Analysis of WTCCC Bipolar Disorder and Control Data The minimum p value from an allelic trend test and three genotypic tests (for dominant, overdominant, and recessive models) is calculated for each marker for CHIAMO and BEAGLE genotype calls. The p values from CHIAMO calls and BEAGLE calls are plotted with the use of a log scale for all markers with minimum p value < 0.0001 for one or both genotype-calling methods. p values for markers that were excluded by data QC filters for CHIAMO calls but not by those for BEAGLE calls are plotted along the line y = 1. p values for markers that were excluded by data QC filters for BEAGLE calls but not by those for CHIAMO calls are plotted along the line x = 1.
Similar articles
- Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500 K array set using 270 HapMap samples.
Hong H, Su Z, Ge W, Shi L, Perkins R, Fang H, Xu J, Chen JJ, Han T, Kaput J, Fuscoe JC, Tong W. Hong H, et al. BMC Bioinformatics. 2008 Aug 12;9 Suppl 9(Suppl 9):S17. doi: 10.1186/1471-2105-9-S9-S17. BMC Bioinformatics. 2008. PMID: 18793462 Free PMC article. - Variability in GWAS analysis: the impact of genotype calling algorithm inconsistencies.
Miclaus K, Chierici M, Lambert C, Zhang L, Vega S, Hong H, Yin S, Furlanello C, Wolfinger R, Goodsaid F. Miclaus K, et al. Pharmacogenomics J. 2010 Aug;10(4):324-35. doi: 10.1038/tpj.2010.46. Pharmacogenomics J. 2010. PMID: 20676070 - Fast two-stage phasing of large-scale sequence data.
Browning BL, Tian X, Zhou Y, Browning SR. Browning BL, et al. Am J Hum Genet. 2021 Oct 7;108(10):1880-1890. doi: 10.1016/j.ajhg.2021.08.005. Epub 2021 Sep 2. Am J Hum Genet. 2021. PMID: 34478634 Free PMC article. - Missing data imputation and haplotype phase inference for genome-wide association studies.
Browning SR. Browning SR. Hum Genet. 2008 Dec;124(5):439-50. doi: 10.1007/s00439-008-0568-7. Epub 2008 Oct 11. Hum Genet. 2008. PMID: 18850115 Free PMC article. Review. - Genotype Imputation in Genome-Wide Association Studies.
Naj AC. Naj AC. Curr Protoc Hum Genet. 2019 Jun;102(1):e84. doi: 10.1002/cphg.84. Curr Protoc Hum Genet. 2019. PMID: 31216114 Review.
Cited by
- Correcting for Sample Contamination in Genotype Calling of DNA Sequence Data.
Flickinger M, Jun G, Abecasis GR, Boehnke M, Kang HM. Flickinger M, et al. Am J Hum Genet. 2015 Aug 6;97(2):284-90. doi: 10.1016/j.ajhg.2015.07.002. Epub 2015 Jul 30. Am J Hum Genet. 2015. PMID: 26235984 Free PMC article. - The paternal and maternal genetic history of Vietnamese populations.
Macholdt E, Arias L, Duong NT, Ton ND, Van Phong N, Schröder R, Pakendorf B, Van Hai N, Stoneking M. Macholdt E, et al. Eur J Hum Genet. 2020 May;28(5):636-645. doi: 10.1038/s41431-019-0557-4. Epub 2019 Dec 11. Eur J Hum Genet. 2020. PMID: 31827276 Free PMC article. - Molecular validation of the schizophrenia spectrum.
Bigdeli TB, Bacanu SA, Webb BT, Walsh D, O'Neill FA, Fanous AH, Riley BP, Kendler KS. Bigdeli TB, et al. Schizophr Bull. 2014 Jan;40(1):60-5. doi: 10.1093/schbul/sbt122. Epub 2013 Aug 22. Schizophr Bull. 2014. PMID: 23970557 Free PMC article. - Characterizing bias in population genetic inferences from low-coverage sequencing data.
Han E, Sinsheimer JS, Novembre J. Han E, et al. Mol Biol Evol. 2014 Mar;31(3):723-35. doi: 10.1093/molbev/mst229. Epub 2013 Nov 27. Mol Biol Evol. 2014. PMID: 24288159 Free PMC article. - The genome and diet of a 35,000-year-old Canis lupus specimen from the Paleolithic painted cave, Chauvet-Pont d'Arc, France.
Elalouf JM, Palacio P, Bon C, Berthonaud V, Maksud F, Stafford TW Jr, Hitte C. Elalouf JM, et al. Ecol Evol. 2022 Aug 23;12(8):e9238. doi: 10.1002/ece3.9238. eCollection 2022 Aug. Ecol Evol. 2022. PMID: 37265549 Free PMC article.
References
- Frayling T.M. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 2007;8:657–662. - PubMed
- Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. - PMC - PubMed
- Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous