Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome - PubMed (original) (raw)
. 2009 Dec 4;326(5958):1369-73.
doi: 10.1126/science.1178535. Epub 2009 Oct 29.
Shashi Bhushan, Alexander Jarasch, Jean-Paul Armache, Soledad Funes, Fabrice Jossinet, James Gumbart, Thorsten Mielke, Otto Berninghausen, Klaus Schulten, Eric Westhof, Reid Gilmore, Elisabet C Mandon, Roland Beckmann
Affiliations
- PMID: 19933108
- PMCID: PMC2920595
- DOI: 10.1126/science.1178535
Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome
Thomas Becker et al. Science. 2009.
Abstract
The trimeric Sec61/SecY complex is a protein-conducting channel (PCC) for secretory and membrane proteins. Although Sec complexes can form oligomers, it has been suggested that a single copy may serve as an active PCC. We determined subnanometer-resolution cryo-electron microscopy structures of eukaryotic ribosome-Sec61 complexes. In combination with biochemical data, we found that in both idle and active states, the Sec complex is not oligomeric and interacts mainly via two cytoplasmic loops with the universal ribosomal adaptor site. In the active state, the ribosomal tunnel and a central pore of the monomeric PCC were occupied by the nascent chain, contacting loop 6 of the Sec complex. This provides a structural basis for the activity of a solitary Sec complex in cotranslational protein translocation.
Figures
Fig. 1. Cryo-EM reconstructions of 80S ribosome-Ssh1 complexes
Cryo-EM reconstructions of the idle (A) and the active (B) 80S-Ssh1 complex at 9 Å resolution. (C) Map of the 80S ribosome with ES27 in the exit conformation at 8 Å. Color code: 40S subunit, yellow; 60S subunit, blue; P-site tRNA/nascent polypeptide chain, green; Ssh1 complexes (PCC), red.
Fig. 2. Visualization of the PCC pore, the nascent polypeptide chain and molecular model
Cut density for the 60S subunit and the idle (A) and active (B) Ssh1 complex is shown as in Fig. 1A. (C) as (A) except ES27 in exit position (dark blue) and P-site tRNA are shown. (D) bottom view of the 6.1 Å RNC map as in Fig. 1. Density for rRNA and ribosomal proteins is highlighted. Asterisk indicates tunnel exit. (E) as (D) showing molecular models. (F) left: isolated density for the P-site tRNA and the nascent DP120 chain (NC) as in (C); right: molecular model for the yeast P-site tRNAAsp and for the nascent DP120 peptide with ribosomal proteins (E).
Fig. 3. A monomeric ribosome-bound Ssh1 complex
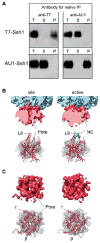
(A) Native immunoprecipitation (IP) of epitope tagged Ssh1 complexes. Microsomes from yeast cells expressing T7 and AU1 tagged Ssh1p were repopulated with RNCs, solubilized, and subjected to native immunoprecipitation using anti-T7 or anti- AU1 antibodies. Total extract (T), supernatant (S) and immunoprecipitate (P) fractions were analyzed by immuno blot using an anti-T7-goat or an anti-AU1-rabbit antibody to detect T7-Ssh1 and AU1-Ssh1. (B) Top: Close-up side views on idle (left) and active (right) PCC (as in Fig. 2A and B). Bottom: Homology models of a monomeric Ssh1 complex (red) fitted into the densities of idle and active Ssh1 complexes (transparent mesh); The cytosolic loop L8 of Ssh1p (red), Sbh1p (β, dark red) and Sss1p (γ, magenta) are indicated. The nascent chain (NC) is shown in green. (C) as (B) showing top views.
Fig. 4. Interaction of the PCC with the ribosome and the nascent chain
(A-C) Molecular models for rRNA and ribosomal proteins are shown as in Fig. 2 E, for the Ssh1 complex as in Fig 3. Views focus on the cytosolic half of the Ssh1 model (A) and the cytosolic loops L6, L8 and the C-terminus (B). (C) A close-up on interactions of cytosolic loops L6 and L8. The positions of the conserved R278 and R411 are indicated (green). (D) Purified Ssh1 complexes from wild type and L6 and L8 mutants were incubated in the presence or absence of yeast ribosomes before centrifugation yielding supernatant (S) and pellet (P) fractions. After SDS-PAGE, Sbh2p was detected using anti-FLAG antibodies.
Fig. 5. The RNC-bound mammalian Sec61 complex is a monomer surrounded by a micelle
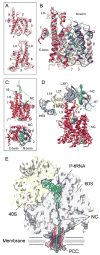
(A) Cryo-EM reconstruction of the 80S RNC-Sec61 complex at 6.5 Å resolution. Top: side view, bottom: bottom view. (B) Isolated density for the Sec61 complex low-pass filtered at 12 Å (top) and at 22 Å (bottom). Connections C1- C4 are indicated. (C) Side view of the ribosome-bound Sec61 complex. The Sec61 model is shown in red, models for ribosomal proteins and rRNA as in Fig. 2 D. The monomeric Sec61 complex fitted into the central portion of the density surrounded by a rim of extra density representing mixed detergent/lipid micelle. (D) Upper left section: side view of the cut densities for the Sec61 complex (red), the surrounding mixed micelle (grey) and the nascent DP120 polypeptide chain and/or the signal anchor sequence (green). Right: Schematic drawing of the mixed micelle of phospholipids (grey) and detergent molecules (blue) surrounding the PCC (red ribbons). Middle section: Isolated densities and schematic drawing as in upper section in a top view (left) or sliced within the plane of the membrane (right). Lower section: Sliced top views, represented as in middle section (left) or as red ribbons for the Sec61 model and transparent mesh for the electron density (right).
Fig. 6. Conformation and nascent polypeptide chain-interactions of the RNC-bound mammalian Sec61 complex
(A) Fit of the Sec61 model (red ribbons) into the density (grey transparent mesh). Side views on the lateral gate (top) and on the cytosolic loops L6 and L8 (bottom). (B) Crystal structure of the M. jannaschii SecYEβ complex (grey) (13) superimposed on Sec61 model. The C- and N-terminal halves are shown in red and blue, transmembrane (TM) helix 7 in yellow and TM helix 2 in cyan. β- (dark red) and γ-subunits (SecE, magenta) are indicated. (C) Side view (top, as in A) and top view (bottom, as in B) of the Sec61 model and extra density for the nascent polypeptide chain (green). (D) Side view as in (C) (top) but rotated to focus on the nascent chain (green). Color code as in Fig. 2E. (E) Schematic representation of an actively translating and translocating eukaryotic ribosome-Sec61 complex with a single copy acting as PCC.
Comment in
- Biochemistry. Nascent proteins caught in the act.
Kampmann M, Blobel G. Kampmann M, et al. Science. 2009 Dec 4;326(5958):1352-3. doi: 10.1126/science.1183690. Science. 2009. PMID: 19965743 No abstract available.
Similar articles
- Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Gogala M, Becker T, Beatrix B, Armache JP, Barrio-Garcia C, Berninghausen O, Beckmann R. Gogala M, et al. Nature. 2014 Feb 6;506(7486):107-10. doi: 10.1038/nature12950. Nature. 2014. PMID: 24499919 - Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex.
Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Beckmann R, et al. Science. 1997 Dec 19;278(5346):2123-6. doi: 10.1126/science.278.5346.2123. Science. 1997. PMID: 9405348 - Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution.
Voorhees RM, Fernández IS, Scheres SH, Hegde RS. Voorhees RM, et al. Cell. 2014 Jun 19;157(7):1632-43. doi: 10.1016/j.cell.2014.05.024. Epub 2014 Jun 12. Cell. 2014. PMID: 24930395 Free PMC article. - Protein translocation by the Sec61/SecY channel.
Osborne AR, Rapoport TA, van den Berg B. Osborne AR, et al. Annu Rev Cell Dev Biol. 2005;21:529-50. doi: 10.1146/annurev.cellbio.21.012704.133214. Annu Rev Cell Dev Biol. 2005. PMID: 16212506 Review. - Structural insight into the protein translocation channel.
Clemons WM Jr, Ménétret JF, Akey CW, Rapoport TA. Clemons WM Jr, et al. Curr Opin Struct Biol. 2004 Aug;14(4):390-6. doi: 10.1016/j.sbi.2004.07.006. Curr Opin Struct Biol. 2004. PMID: 15313231 Review.
Cited by
- The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum.
Tirincsi A, Sicking M, Hadzibeganovic D, Haßdenteufel S, Lang S. Tirincsi A, et al. Int J Mol Sci. 2021 Dec 23;23(1):143. doi: 10.3390/ijms23010143. Int J Mol Sci. 2021. PMID: 35008565 Free PMC article. Review. - Molecular mechanism of GTPase activation at the signal recognition particle (SRP) RNA distal end.
Shen K, Wang Y, Hwang Fu YH, Zhang Q, Feigon J, Shan SO. Shen K, et al. J Biol Chem. 2013 Dec 20;288(51):36385-97. doi: 10.1074/jbc.M113.513614. Epub 2013 Oct 22. J Biol Chem. 2013. PMID: 24151069 Free PMC article. - Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems.
Brodsky JL, Skach WR. Brodsky JL, et al. Curr Opin Cell Biol. 2011 Aug;23(4):464-75. doi: 10.1016/j.ceb.2011.05.004. Epub 2011 Jun 12. Curr Opin Cell Biol. 2011. PMID: 21664808 Free PMC article. Review. - MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments.
Wild K, Aleksić M, Lapouge K, Juaire KD, Flemming D, Pfeffer S, Sinning I. Wild K, et al. Nat Commun. 2020 Feb 7;11(1):776. doi: 10.1038/s41467-020-14603-7. Nat Commun. 2020. PMID: 32034140 Free PMC article. - Single-particle electron microscopy in the study of membrane protein structure.
De Zorzi R, Mi W, Liao M, Walz T. De Zorzi R, et al. Microscopy (Oxf). 2016 Feb;65(1):81-96. doi: 10.1093/jmicro/dfv058. Epub 2015 Oct 15. Microscopy (Oxf). 2016. PMID: 26470917 Free PMC article.
References
- Rapoport TA. Nature. 2007;450:663. - PubMed
- Halic M, Beckmann R. Curr Opin Struct Biol. 2005;15:116. - PubMed
- Beckmann R, et al. Science. 1997;278:2123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-GM067887/GM/NIGMS NIH HHS/United States
- P41 RR005969/RR/NCRR NIH HHS/United States
- R01 GM067887/GM/NIGMS NIH HHS/United States
- GM35687/GM/NIGMS NIH HHS/United States
- R01 GM035687/GM/NIGMS NIH HHS/United States
- P41-RR05969/RR/NCRR NIH HHS/United States
- P41 RR005969-19/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases