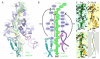Structural insight into nascent polypeptide chain-mediated translational stalling - PubMed (original) (raw)
. 2009 Dec 4;326(5958):1412-5.
doi: 10.1126/science.1177662. Epub 2009 Oct 29.
C Axel Innis, Daniel N Wilson, Marco Gartmann, Jean-Paul Armache, Elizabeth Villa, Leonardo G Trabuco, Thomas Becker, Thorsten Mielke, Klaus Schulten, Thomas A Steitz, Roland Beckmann
Affiliations
- PMID: 19933110
- PMCID: PMC2920484
- DOI: 10.1126/science.1177662
Structural insight into nascent polypeptide chain-mediated translational stalling
Birgit Seidelt et al. Science. 2009.
Abstract
Expression of the Escherichia coli tryptophanase operon depends on ribosome stalling during translation of the upstream TnaC leader peptide, a process for which interactions between the TnaC nascent chain and the ribosomal exit tunnel are critical. We determined a 5.8 angstrom-resolution cryo-electron microscopy and single-particle reconstruction of a ribosome stalled during translation of the tnaC leader gene. The nascent chain was extended within the exit tunnel, making contacts with ribosomal components at distinct sites. Upon stalling, two conserved residues within the peptidyltransferase center adopted conformations that preclude binding of release factors. We propose a model whereby interactions within the tunnel are relayed to the peptidyltransferase center to inhibit translation. Moreover, we show that nascent chains adopt distinct conformations within the ribosomal exit tunnel.
Figures
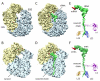
Fig. 1
Cryo-EM reconstruction of the TnaC•70S complex. (A-B) Cryo-EM reconstruction of the control E. coli 70S ribosome at 6.6 Å resolution, with small and large subunit colored yellow and blue, respectively. (C-D) The 5.8 Å resolution cryo-EM density of the TnaC•70S complex, with density for the TnaC-tRNA shown in green. (E) Isolated density for the TnaCtRNA (green) and mRNA (red) from (C). The relative positions of ribosomal proteins L4 (purple), L22 (blue) and L23 (yellow) are indicated. (F) Fitting of molecular models for the TnaC-tRNAPro into the cryo-EM density from (E).
Fig. 2
Interaction between the TnaC nascent chain and the ribosomal tunnel. The central panel shows a section through the large subunit revealing contact points between the TnaC nascent chain (light green) and the ribosome (gray). Density attributed to the P-tRNA is coloured dark green, and for peptidyl-tRNA at a lower threshold is shown as a green mesh. (A)-(D) Different views of connections between the nascent chain (green ribbon) and the 23S rRNA (blue sticks) or ribosomal protein L22 (blue ribbon). Density for the TnaC•70S complex is shown as a transparent gray surface, whereas the isolated nascent chain at a lower threshold is shown as a green mesh. Important TnaC residues are highlighted yellow, orange or red.
Fig. 3
Silencing of the peptidyltransferase center (PTC). (A) Conformation of 23S rRNA nucleotides at the PTC when tRNA CCA-end mimics are bound to A- (cyan) and P-sites (green) (PDB1VQN). (B) Comparison of A2602 and U2585 conformations in various ribosome crystal structures (red, PDB1VQK; teal, PDB1S72; yellow, PDB2I2T; gold, PDB2JL5/6; blue, PDB1VQ9; green, PDB1VQN). (C) View into the PTC of the TnaC-70S complex, with the MDFF model of the TnaC-tRNA (green) and nucleotides of the 23S rRNA (blue). The cryo-EM density is shown as a transparent gray surface, with an asterisk indicating the connection between P-tRNA and nascent chain. (D) View into the PTC of 70SRNC complex, with fitted models as in (A). Note the lack of density (gray) for nucleotide A2602. (E) As in (A), but with the antibiotic sparsomycin (SPAR, red; PDB1VQ9) and the terminal A76 and aminoacyl moiety of an A-tRNA (cyan; PDB1VQN) included. (F) Comparison of A2602 and U2585 positions (arrowed) between TnaC-70S complex (blue) and RF2-70S complex (24); gold), with RF2 shown as surface representation (gold).
Fig. 4
Relay for PTC silencing and the path of the nascent chain. (A) Ribosomal components potentially involved in a relay mechanism to inactivate the PTC, with those implicated in stalling in bold. The TnaC nascent chain is in green, with residues essential for stalling colored yellow. The isolated TnaC-tRNA density is shown as a transparent gray surface. (B) Schematic indicating potential relay pathways from Trp12 (W12) of TnaC to the PTC, either through the nascent chain itself (R1) or via networks of interconnected 23S rRNA nucleotides (R2 and R3). (C-D) Transverse section through the large subunit showing the path of (C) TnaC (dark green) and (D) DP120 (35); orange) nascent chains through the ribosomal tunnel. (E) Superposition of (A) and (B). (F) Schematic highlighting the similarities and differences between the TnaC and DP120 nascent chain in terms of contacts and passage through the tunnel.
Comment in
- Biochemistry. Nascent proteins caught in the act.
Kampmann M, Blobel G. Kampmann M, et al. Science. 2009 Dec 4;326(5958):1352-3. doi: 10.1126/science.1183690. Science. 2009. PMID: 19965743 No abstract available.
Similar articles
- 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction.
Yang R, Cruz-Vera LR, Yanofsky C. Yang R, et al. J Bacteriol. 2009 Jun;191(11):3445-50. doi: 10.1128/JB.00096-09. Epub 2009 Mar 27. J Bacteriol. 2009. PMID: 19329641 Free PMC article. - Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression.
Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Cruz-Vera LR, et al. Mol Cell. 2005 Aug 5;19(3):333-43. doi: 10.1016/j.molcel.2005.06.013. Mol Cell. 2005. PMID: 16061180 - Modulating the activity of the peptidyl transferase center of the ribosome.
Beringer M. Beringer M. RNA. 2008 May;14(5):795-801. doi: 10.1261/rna.980308. Epub 2008 Mar 27. RNA. 2008. PMID: 18369182 Free PMC article. Review. - Ribosomal tunnel and translation regulation.
Bogdanov AA, Sumbatyan NV, Shishkina AV, Karpenko VV, Korshunova GA. Bogdanov AA, et al. Biochemistry (Mosc). 2010 Dec;75(13):1501-16. doi: 10.1134/s0006297910130018. Biochemistry (Mosc). 2010. PMID: 21417991 Review.
Cited by
- Exploiting translational stalling peptides in an effort to extend azithromycin interaction within the prokaryotic ribosome nascent peptide exit tunnel.
Washington AZ, Tapadar S, George A, Oyelere AK. Washington AZ, et al. Bioorg Med Chem. 2015 Aug 15;23(16):5198-209. doi: 10.1016/j.bmc.2015.04.078. Epub 2015 May 6. Bioorg Med Chem. 2015. PMID: 26037612 Free PMC article. - Force transduction creates long-ranged coupling in ribosomes stalled by arrest peptides.
Zimmer MH, Niesen MJM, Miller TF 3rd. Zimmer MH, et al. Biophys J. 2021 Jun 15;120(12):2425-2435. doi: 10.1016/j.bpj.2021.03.041. Epub 2021 Apr 29. Biophys J. 2021. PMID: 33932440 Free PMC article. - Rock, scissors, paper: How RNA structure informs function.
Assmann SM, Chou HL, Bevilacqua PC. Assmann SM, et al. Plant Cell. 2023 May 29;35(6):1671-1707. doi: 10.1093/plcell/koad026. Plant Cell. 2023. PMID: 36747354 Free PMC article. - Proline-Rich Antimicrobial Peptides in Medicinal Maggots of Lucilia sericata Interact With Bacterial DnaK But Do Not Inhibit Protein Synthesis.
Cytryńska M, Rahnamaeian M, Zdybicka-Barabas A, Dobslaff K, Züchner T, Sacheau G, Innis CA, Vilcinskas A. Cytryńska M, et al. Front Pharmacol. 2020 Apr 24;11:532. doi: 10.3389/fphar.2020.00532. eCollection 2020. Front Pharmacol. 2020. PMID: 32390853 Free PMC article. - Universal and domain-specific sequences in 23S-28S ribosomal RNA identified by computational phylogenetics.
Doris SM, Smith DR, Beamesderfer JN, Raphael BJ, Nathanson JA, Gerbi SA. Doris SM, et al. RNA. 2015 Oct;21(10):1719-30. doi: 10.1261/rna.051144.115. Epub 2015 Aug 17. RNA. 2015. PMID: 26283689 Free PMC article.
References
- Woolhead CA, McCormick PJ, Johnson AE. Cell. 2004;116:725–36. - PubMed
- Lu J, Deutsch C. Biochemistry. 2005;44:8230–43. - PubMed
- Lu J, Deutsch C. Nat Struct Mol Biol. 2005;12:1123–9. - PubMed
- Tenson T, Ehrenberg M. Cell. 2002;108:591–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR005969/RR/NCRR NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P01 GM022778/GM/NIGMS NIH HHS/United States
- GM022778/GM/NIGMS NIH HHS/United States
- P41-RR05969/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases