The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway - PubMed (original) (raw)
The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway
Jana Rudolf et al. Nucleic Acids Res. 2010 Jan.
Abstract
Xeroderma pigmentosum factor D (XPD) is a 5'-3' superfamily 2 helicase and the founding member of a family of DNA helicases with iron-sulphur cluster domains. As a component of transcription factor II H (TFIIH), XPD is involved in DNA unwinding during nucleotide excision repair (NER). Archaeal XPD is closely related in sequence to the eukaryal enzyme and the crystal structure of the archaeal enzyme has provided a molecular understanding of mutations causing xeroderma pigmentosum and trichothiodystrophy in humans. Consistent with a role in NER, we show that archaeal XPD can initiate unwinding from a DNA bubble structure, differentiating it from the related helicases FancJ and DinG. XPD was not stalled by substrates containing extrahelical fluorescein adducts, abasic sites nor a cyclobutane pyrimidine dimer, regardless of whether these modifications were placed on either the displaced or translocated strands. This suggests that DNA lesions repaired by NER may not present a barrier to XPD translocation in vivo, in contrast to some predictions. Preferential binding of a fluorescein-adducted oligonucleotide was observed, and XPD helicase activity was readily inhibited by both single- and double-stranded DNA binding proteins. These observations have several implications for the current understanding of the NER pathway.
Figures
Figure 1.
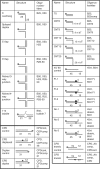
DNA constructs used in this study. Substrates were prepared by annealing the oligonucleotides indicated followed by purification from native polyacrylamide gels. The position of the 5′-32P-label is indicated by a grey circle. The positions of modifications are indicated with symbols and discussed in the text.
Figure 2.
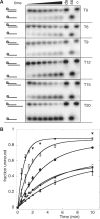
Substrate requirements of XPD. The DNA unwinding activity of XPD increases with increasing length of the 5′ ssDNA overhang. (A) Representative 12% acrylamide:TBE gels of XPD (200 nM) unwinding DNA providing 0, 6, 9, 12, 15 and 20 nt of 5′ ssDNA dT-overhangs (T0–T20) at 45°C. Time points were 0.5, 1, 1.5, 2, 3, 5 and 10 min. Controls: ss, boiled substrate; ds, intact substrate; C, substrate stability in the absence of XPD at the end point of the reaction. (B) The graph shows quantification of results of triplicate experiments with the standard errors indicated. The fraction of total DNA unwinding is plotted against the time and data points were fitted using equation 1. T0 (filled triangle); T6 (open square); T9 (filled diamond); T12 (filled circle); T15 (open circle) and T20 (inverted filled trianlge).
Figure 3.
ITC analysis of DNA binding affinity. Isothermal titration calorimetry profiles for the interaction of XPD with oligonucleotides of different lengths. The top panel shows heat differences obtained for injections of 148 µM (ITC-T8 and ITC-T12) and 113 µM (ITC-T16 and ITC-T16F) XPD into 5 µM DNA solutions. The lower panel shows the incremental enthalpy changes, corrected for heats of dilution, with experimental points (open square) and the best fit (thin line). Data were fitted using a one-site binding model.
Figure 4.
DNA substrate preference of XPD. The figure shows representative 12% acrylamide:TBE gels. The DNA structures before and after unwinding are shown to the left of the gels. Time points were 0.5, 1, 1.5, 2, 3, 5 and 10 min for all substrates except for the nicked three-way and four-way junctions (5, 10, 20 and 30 min). Controls: ss, boiled substrate; ds, intact substrate; C, substrate stability in the absence of XPD at the end point of the reaction. (A) XPD (100 nM) unwinding a 5′ overhang, splayed duplex and 5′ flap DNA substrate at 45°C. Reaction rates were all within a factor of 2. (B) XPD (500 nM) unwinding a 5′ overhang, a 3′ flap, a nicked three-way and a nicked four-way junction at 35°C. No significant unwinding was observed. (C) XPD (200 nM) unwinding a 7 nt bubble substrate at 45°C. The plot shows quantification of triplicate experiments with means and standard errors shown for the 7 nt bubble (filled circle) and a 25 nt overhang substrate (open circle).
Figure 5.
DNA modifications on the displaced or translocated strands do not affect the helicase activity of XPD. Representative 12% acrylamide:TBE gels are shown. Time points were 0.5, 1, 1.5, 2, 3, 5 and 10 min at 45°C for all experiments except those shown in the bottom panel where the CPD was in the translocating strand, where time points were 0.2, 1, 2, 5, 10, 15 and 20 min at 30°C. The DNA structures used are described in Figure 1. Filled squares represent the fluorescein backbone modification (A), open squares the position of the abasic site (B) and triangles the position of the CPD lesion (C). Controls were as follows: ss, boiled substrate; ds, intact substrate; C, substrate stability in the absence of ATP at the end point of the reaction; C88S, substrates incubated with an inactive mutant of XPD. The plots on the right show means and standard error bars for triplicate experiments, fitted using Equation 1, with the following symbols: (open circle), control, no modification; (filled circle), displaced strand; (open square), translocated strand; (filled diamond), both strands.
Figure 6.
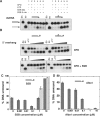
The influence of DNA binding proteins on the helicase activity of XPD. (A) DNA helicase assays were carried out using 10 nM of a 31 nt dsDNA substrate with or without a CPD lesion (Figure 1). The reactions were incubated at 45°C using the indicated combinations of 250 nM XPD, 500 nM SSB, 0.5 mM Mg-ATP. Controls are ds: DNA substrate; ss: boiled DNA. (B) Representative gels showing time courses of DNA unwinding reactions using a 31 nt duplex DNA substrate with or without a CPD lesion flanked by a 15 nt 5′overhang. Experiments were carried out with 250 nM XPD in the absence of SSB for 1, 2, 5, 10 and 15 min (top) or presence of 250 nM SSB for 0.5, 1, 2, 5 and 10 min (bottom). Controls are ds: DNA substrate; ss: boiled DNA; –ATP: substrate stability in the absence of ATP at the end point of the reaction. (C) The influence of increasing concentrations of SSB on the helicase activity of XPD is illustrated. The plot shows the mean values of duplicate experiments with the standard errors indicated. Experiments were carried out in the presence (dark grey bars) and absence (light grey bars) of 200 nM XPD at 45°C using the 45/60 control DNA substrate (10 nM, Figure 1) and increasing concentrations of SSB for 3 min. (D) The influence of increasing concentrations of Alba1 on the helicase activity of XPD is illustrated. For experimental conditions and analysis see above.
Figure 7.
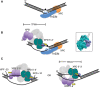
Model for the early steps in the eukaryal nucleotide excision repair pathway. (A) Initial damage recognition is carried out by hr23B-XPC, which binds the undamaged strand opposite a lesion as well as the duplex DNA downstream, opening a small ssDNA bubble. (B) Binding of TFIIH results in opening of at least 10 bp upstream (5′) of the damage site—enough to accommodate XPD on the damaged strand but not enough space to bind XPB simultaneously. XPD could extend the bubble by unwinding DNA in a 5′ to 3′ direction. The inset shows the structure of XPD from Thermoplasma acidophilum with the likely path of the DNA indicated. (C) Once the ssDNA bubble has been extended, XPB could bind at the 5′-end. The NER patch size is likely to be defined by the length of DNA bound by the combined actions of XPD and XPB. XPD can unwind DNA past the lesion but could be stalled by collision with hr23B-XPC and/or by constraints imposed by the rest of the TFIIH complex including XPB. In subsequent steps XPA and RPA replace the hr23B-XPC complex, and cleavage is catalysed by XPG and XPF-ERCC1. Alternatively, XPB could bind on the undamaged strand. Both helicases would then migrate in the same direction, tracking on separate strands.
Similar articles
- XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. Fan L, et al. Cell. 2008 May 30;133(5):789-800. doi: 10.1016/j.cell.2008.04.030. Cell. 2008. PMID: 18510924 Free PMC article. - XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase.
Fuss JO, Tainer JA. Fuss JO, et al. DNA Repair (Amst). 2011 Jul 15;10(7):697-713. doi: 10.1016/j.dnarep.2011.04.028. Epub 2011 May 14. DNA Repair (Amst). 2011. PMID: 21571596 Free PMC article. Review. - The DNA repair helicases XPD and FancJ have essential iron-sulfur domains.
Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. Rudolf J, et al. Mol Cell. 2006 Sep 15;23(6):801-8. doi: 10.1016/j.molcel.2006.07.019. Mol Cell. 2006. PMID: 16973432 - Structure, function and evolution of the XPD family of iron-sulfur-containing 5'-->3' DNA helicases.
White MF. White MF. Biochem Soc Trans. 2009 Jun;37(Pt 3):547-51. doi: 10.1042/BST0370547. Biochem Soc Trans. 2009. PMID: 19442249 - The XPD helicase: XPanDing archaeal XPD structures to get a grip on human DNA repair.
Wolski SC, Kuper J, Kisker C. Wolski SC, et al. Biol Chem. 2010 Jul;391(7):761-5. doi: 10.1515/BC.2010.076. Biol Chem. 2010. PMID: 20482310 Review.
Cited by
- Impact of age-associated cyclopurine lesions on DNA repair helicases.
Khan I, Suhasini AN, Banerjee T, Sommers JA, Kaplan DL, Kuper J, Kisker C, Brosh RM Jr. Khan I, et al. PLoS One. 2014 Nov 19;9(11):e113293. doi: 10.1371/journal.pone.0113293. eCollection 2014. PLoS One. 2014. PMID: 25409515 Free PMC article. - Taking a molecular motor for a spin: helicase mechanism studied by spin labeling and PELDOR.
Constantinescu-Aruxandei D, Petrovic-Stojanovska B, Schiemann O, Naismith JH, White MF. Constantinescu-Aruxandei D, et al. Nucleic Acids Res. 2016 Jan 29;44(2):954-68. doi: 10.1093/nar/gkv1373. Epub 2015 Dec 10. Nucleic Acids Res. 2016. PMID: 26657627 Free PMC article. - Mechanism of DNA loading by the DNA repair helicase XPD.
Constantinescu-Aruxandei D, Petrovic-Stojanovska B, Penedo JC, White MF, Naismith JH. Constantinescu-Aruxandei D, et al. Nucleic Acids Res. 2016 Apr 7;44(6):2806-15. doi: 10.1093/nar/gkw102. Epub 2016 Feb 20. Nucleic Acids Res. 2016. PMID: 26896802 Free PMC article. - Close encounters for the first time: Helicase interactions with DNA damage.
Khan I, Sommers JA, Brosh RM Jr. Khan I, et al. DNA Repair (Amst). 2015 Sep;33:43-59. doi: 10.1016/j.dnarep.2015.06.003. Epub 2015 Jun 16. DNA Repair (Amst). 2015. PMID: 26160335 Free PMC article. Review. - Subunit architecture of general transcription factor TFIIH.
Gibbons BJ, Brignole EJ, Azubel M, Murakami K, Voss NR, Bushnell DA, Asturias FJ, Kornberg RD. Gibbons BJ, et al. Proc Natl Acad Sci U S A. 2012 Feb 7;109(6):1949-54. doi: 10.1073/pnas.1105266109. Epub 2012 Jan 20. Proc Natl Acad Sci U S A. 2012. PMID: 22308316 Free PMC article.
References
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429.
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. - PubMed
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. - PubMed
- Feaver WJ, Svejstrup JQ, Bardwell L, Bardwell AJ, Buratowski S, Gulyas KD, Donahue TF, Friedberg EC, Kornberg RD. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous