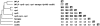Pseudomonas aeruginosa population structure revisited - PubMed (original) (raw)
Pseudomonas aeruginosa population structure revisited
Jean-Paul Pirnay et al. PLoS One. 2009.
Abstract
At present there are strong indications that Pseudomonas aeruginosa exhibits an epidemic population structure; clinical isolates are indistinguishable from environmental isolates, and they do not exhibit a specific (disease) habitat selection. However, some important issues, such as the worldwide emergence of highly transmissible P. aeruginosa clones among cystic fibrosis (CF) patients and the spread and persistence of multidrug resistant (MDR) strains in hospital wards with high antibiotic pressure, remain contentious. To further investigate the population structure of P. aeruginosa, eight parameters were analyzed and combined for 328 unrelated isolates, collected over the last 125 years from 69 localities in 30 countries on five continents, from diverse clinical (human and animal) and environmental habitats. The analysed parameters were: i) O serotype, ii) Fluorescent Amplified-Fragment Length Polymorphism (FALFP) pattern, nucleotide sequences of outer membrane protein genes, iii) oprI, iv) oprL, v) oprD, vi) pyoverdine receptor gene profile (fpvA type and fpvB prevalence), and prevalence of vii) exoenzyme genes exoS and exoU and viii) group I pilin glycosyltransferase gene tfpO. These traits were combined and analysed using biological data analysis software and visualized in the form of a minimum spanning tree (MST). We revealed a network of relationships between all analyzed parameters and non-congruence between experiments. At the same time we observed several conserved clones, characterized by an almost identical data set. These observations confirm the nonclonal epidemic population structure of P. aeruginosa, a superficially clonal structure with frequent recombinations, in which occasionally highly successful epidemic clones arise. One of these clones is the renown and widespread MDR serotype O12 clone. On the other hand, we found no evidence for a widespread CF transmissible clone. All but one of the 43 analysed CF strains belonged to a ubiquitous P. aeruginosa "core lineage" and typically exhibited the exoS(+)/exoU(-) genotype and group B oprL and oprD alleles. This is to our knowledge the first report of an MST analysis conducted on a polyphasic data set.
Conflict of interest statement
Competing Interests: Dr. Pot is affiliated with Applied Maths. Applied Maths played no role in the funding of the research.
Figures
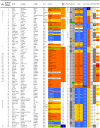
Figure 1. Overview of the characteristics and test results of P. aeruginosa strains 1-82/328.
ABR: antibiotic resistance; ATCC: American Type Culture Collection; BWC: burn wound centre; CC: clonal complex; CF: cystic fibrosis; COPD: chronic obstructive pulmonary disease; CPHL: Central Public Health Laboratory, London; DOM: defective oprD mutation; ESP: Ecole de Santé Publique, Brussels; FAFLP: fluorescent amplified fragment length polymorphism; HPA: Health protection Agency, Colindale; HSV: high sequence variability; IDEXX: laboratory for veterinary, food and water testing; ICU: intensive care unit; IG: insufficient growth; LMG: Laboratorium voor Microbiologie Gent, public bacteria collection; NA: no amplification; NIMR: National Institute for Medical Research, London; NT: not typable; POS: positive; TRM: reaction terminated. * ABR: antibiotic resistance, expressed as the number of antibiotic classes to which resistance was observed. ** Detected using degenerate primers.
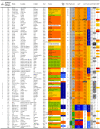
Figure 2. Overview of the characteristics and test results of P. aeruginosa strains 83-164/328.
Figure 3. Overview of the characteristics and test results of P. aeruginosa strains 165-246/328.
Figure 4. Overview of the characteristics and test results of P. aeruginosa strains 247-328/328.
Figure 5. Congruence between experiments, as calculated using the Pearson product-moment correlation coefficient.
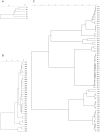
Figure 6. UPGMA dendrograms, with assignment of allele codes, for the oprI (a), oprL (b), and oprD (c) alleles detected in 328 P. aeruginosa strains.
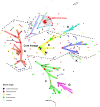
Figure 7. Minimum spanning tree of the similarity matrix of the composite data set consisting of the FAFLP pattern, serotype, oprI, oprL, and oprD gene sequences, pyoverdine receptor profile and prevalence of exoS/U for 328 P. aeruginosa strains.
Each circle corresponds to a polyphasic profile (PP). The circles are scaled with member count. Branch lengths are logarithmic. Coloured zones surround PPs that belong to the same clonal complex. These complexes are also indicated with a capital letter. The lines between PPs indicate inferred phylogenetic relationships and are represented as bold, plain, discontinuous and light discontinuous depending on the number of differences between profile types. Discontinuous links are only indicative. Two bold black indent lines delimit the P. aeruginosa “core lineage”; the MDR serotype O12 clone is encircled by a red dotted line.
Similar articles
- Pseudomonas aeruginosa displays an epidemic population structure.
Pirnay JP, De Vos D, Cochez C, Bilocq F, Vanderkelen A, Zizi M, Ghysels B, Cornelis P. Pirnay JP, et al. Environ Microbiol. 2002 Dec;4(12):898-911. doi: 10.1046/j.1462-2920.2002.00321.x. Environ Microbiol. 2002. PMID: 12534471 - Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales.
Scott FW, Pitt TL. Scott FW, et al. J Med Microbiol. 2004 Jul;53(Pt 7):609-615. doi: 10.1099/jmm.0.45620-0. J Med Microbiol. 2004. PMID: 15184530 - Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone.
Pirnay JP, De Vos D, Cochez C, Bilocq F, Pirson J, Struelens M, Duinslaeger L, Cornelis P, Zizi M, Vanderkelen A. Pirnay JP, et al. J Clin Microbiol. 2003 Mar;41(3):1192-202. doi: 10.1128/JCM.41.3.1192-1202.2003. J Clin Microbiol. 2003. PMID: 12624051 Free PMC article. - Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis.
Parkins MD, Somayaji R, Waters VJ. Parkins MD, et al. Clin Microbiol Rev. 2018 Aug 29;31(4):e00019-18. doi: 10.1128/CMR.00019-18. Print 2018 Oct. Clin Microbiol Rev. 2018. PMID: 30158299 Free PMC article. Review. - The increasing threat of Pseudomonas aeruginosa high-risk clones.
Oliver A, Mulet X, López-Causapé C, Juan C. Oliver A, et al. Drug Resist Updat. 2015 Jul-Aug;21-22:41-59. doi: 10.1016/j.drup.2015.08.002. Epub 2015 Aug 10. Drug Resist Updat. 2015. PMID: 26304792 Review.
Cited by
- Isolation and Characterization of Lytic Pseudomonas aeruginosa Bacteriophages Isolated from Sewage Samples from Tunisia.
Akremi I, Merabishvili M, Jlidi M, Haj Brahim A, Ben Ali M, Karoui A, Lavigne R, Wagemans J, Pirnay JP, Ben Ali M. Akremi I, et al. Viruses. 2022 Oct 25;14(11):2339. doi: 10.3390/v14112339. Viruses. 2022. PMID: 36366441 Free PMC article. - A panel of diverse Pseudomonas aeruginosa clinical isolates for research and development.
Lebreton F, Snesrud E, Hall L, Mills E, Galac M, Stam J, Ong A, Maybank R, Kwak YI, Johnson S, Julius M, Ly M, Swierczewski B, Waterman PE, Hinkle M, Jones A, Lesho E, Bennett JW, McGann P. Lebreton F, et al. JAC Antimicrob Resist. 2021 Dec 10;3(4):dlab179. doi: 10.1093/jacamr/dlab179. eCollection 2021 Dec. JAC Antimicrob Resist. 2021. PMID: 34909689 Free PMC article. - Genome-wide patterns of recombination in the opportunistic human pathogen Pseudomonas aeruginosa.
Dettman JR, Rodrigue N, Kassen R. Dettman JR, et al. Genome Biol Evol. 2014 Dec 4;7(1):18-34. doi: 10.1093/gbe/evu260. Genome Biol Evol. 2014. PMID: 25480685 Free PMC article. - Establishing Genotype-to-Phenotype Relationships in Bacteria Causing Hospital-Acquired Pneumonia: A Prelude to the Application of Clinical Metagenomics.
Ruppé E, Cherkaoui A, Lazarevic V, Emonet S, Schrenzel J. Ruppé E, et al. Antibiotics (Basel). 2017 Nov 29;6(4):30. doi: 10.3390/antibiotics6040030. Antibiotics (Basel). 2017. PMID: 29186015 Free PMC article. Review. - RX-P873, a Novel Protein Synthesis Inhibitor, Accumulates in Human THP-1 Monocytes and Is Active against Intracellular Infections by Gram-Positive (Staphylococcus aureus) and Gram-Negative (Pseudomonas aeruginosa) Bacteria.
Buyck JM, Peyrusson F, Tulkens PM, Van Bambeke F. Buyck JM, et al. Antimicrob Agents Chemother. 2015 Aug;59(8):4750-8. doi: 10.1128/AAC.00428-15. Epub 2015 May 26. Antimicrob Agents Chemother. 2015. PMID: 26014952 Free PMC article.
References
- Gessard C. Sur les colorations bleue et verte des linges à pansements. Comptes-rendus hebdomadaire des séances de l'Académie des Sciences. 1882;94:536–538.
- Goldberg JB. Pseudomonas: global bacteria. Trends Microbiol. 2000;8:55–57. - PubMed
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. - PubMed
- Foght JM, Westlake DWS, Johnson WM, Ridgway HF. Environmental gasoline-utilizing isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology. 1996;142:2333–2340. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources