HIF-1: upstream and downstream of cancer metabolism - PubMed (original) (raw)
Review
HIF-1: upstream and downstream of cancer metabolism
Gregg L Semenza. Curr Opin Genet Dev. 2010 Feb.
Abstract
Hypoxia-inducible factor 1 (HIF-1) plays a key role in the reprogramming of cancer metabolism by activating transcription of genes encoding glucose transporters and glycolytic enzymes, which take up glucose and convert it to lactate; pyruvate dehydrogenase kinase 1, which shunts pyruvate away from the mitochondria; and BNIP3, which triggers selective mitochondrial autophagy. The shift from oxidative to glycolytic metabolism allows maintenance of redox homeostasis and cell survival under conditions of prolonged hypoxia. Many metabolic abnormalities in cancer cells increase HIF-1 activity. As a result, a feed-forward mechanism can be activated that drives HIF-1 activation and may promote tumor progression.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures
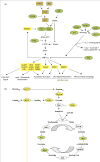
Figure 1
HIF-1 and metabolism. (A) Regulation of HIF-1α protein synthesis and stability and HIF-1-dependent metabolic reprogramming. The rate of translation of HIF-1α mRNA into protein in cancer cells is dependent upon the activity of the mammalian target of rapamycin (mTOR), which is determined by the activity of upstream tumor suppressor proteins (green ovals) and oncoproteins (orange rectangles). Arrows indicate stimulation, blocked lines indicate inhibition. HIF-1α protein stability is regulated by O2- and α-ketoglutarate-dependent prolyl hydroxylation catalyzed by PHD2. Hydroxylation is required for the binding of the von Hippel-Lindau protein (VHL), which recruits a ubiquitin ligase that targets HIF-1α for proteasomal degradation. Loss of function for any of the tumor suppressor genes encoding fumarate hydratase (FH), isocitrate dehydrogenase (IDH), or succinate dehydrogenase (SDH) inhibits PHD2 activity. HIF-1α dimerizes with HIF-1β (not shown) and activates transcription of target genes encoding proteins (yellow rectangles) that play key roles in the metabolic reprogramming of cancer cells. Abbreviations: PTKs, protein tyrosine kinases; PI3K, phosphatidylinositol-3-kinase; S6K, ribosomal protein S6 kinase; ALD, aldolase; PGK, phosphoglycerate kinase; ENO, enolase; PKM, pyruvate kinase M. Other HIF-1-regulated glycolytic enzymes that are not shown: glucosephosphate isomerase, phosphofructokinase L, triosephosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglyceromutase. (B) Oxidative and glycolytic metabolism of glucose. HIF-1-regulated genes (yellow ovals) play key roles in converting extracellular glucose to extracellular lactate and blocking entry of pyruvate into the tricarboxylic acid cycle. Loss of function for each of the enzymes shown in green is associated with tumor formation and HIF-1α stabilization due to PHD2 inhibition by accumulation of the enzyme substrate. Arrows indicate the direction of the cycle. CS, citrate synthase; αKGDH, α-ketoglutarate dehydrogenase; SCS, succinyl-CoA synthetase; MDH, malate dehydrogenase.
Similar articles
- Regulation of metabolism by hypoxia-inducible factor 1.
Semenza GL. Semenza GL. Cold Spring Harb Symp Quant Biol. 2011;76:347-53. doi: 10.1101/sqb.2011.76.010678. Epub 2011 Jul 22. Cold Spring Harb Symp Quant Biol. 2011. PMID: 21785006 Review. - Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells.
Luo W, Semenza GL. Luo W, et al. Oncotarget. 2011 Jul;2(7):551-6. doi: 10.18632/oncotarget.299. Oncotarget. 2011. PMID: 21709315 Free PMC article. - The intravenous anesthetic propofol inhibits lipopolysaccharide-induced hypoxia-inducible factor 1 activation and suppresses the glucose metabolism in macrophages.
Tanaka T, Takabuchi S, Nishi K, Oda S, Wakamatsu T, Daijo H, Fukuda K, Hirota K. Tanaka T, et al. J Anesth. 2010 Feb;24(1):54-60. doi: 10.1007/s00540-009-0829-1. Epub 2009 Dec 29. J Anesth. 2010. PMID: 20039079 - Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning.
Semenza GL. Semenza GL. Biochim Biophys Acta. 2011 Jul;1813(7):1263-8. doi: 10.1016/j.bbamcr.2010.08.006. Epub 2010 Aug 21. Biochim Biophys Acta. 2011. PMID: 20732359 Free PMC article. Review. - Regulation of cancer cell metabolism by hypoxia-inducible factor 1.
Semenza GL. Semenza GL. Semin Cancer Biol. 2009 Feb;19(1):12-6. doi: 10.1016/j.semcancer.2008.11.009. Epub 2008 Dec 9. Semin Cancer Biol. 2009. PMID: 19114105 Review.
Cited by
- Hypoxia, cancer metabolism and the therapeutic benefit of targeting lactate/H(+) symporters.
Marchiq I, Pouysségur J. Marchiq I, et al. J Mol Med (Berl). 2016 Feb;94(2):155-71. doi: 10.1007/s00109-015-1307-x. Epub 2015 Jun 24. J Mol Med (Berl). 2016. PMID: 26099350 Free PMC article. Review. - p16INK4A represses breast stromal fibroblasts migration/invasion and their VEGF-A-dependent promotion of angiogenesis through Akt inhibition.
Al-Ansari MM, Hendrayani SF, Tulbah A, Al-Tweigeri T, Shehata AI, Aboussekhra A. Al-Ansari MM, et al. Neoplasia. 2012 Dec;14(12):1269-77. doi: 10.1593/neo.121632. Neoplasia. 2012. PMID: 23308058 Free PMC article. - Selective enhancement of hypoxic cell killing by tempol-regulated suicide gene expression.
Kagiya G, Ogawa R, Choudhuri R, Cook JA, Hatashita M, Tanaka Y, Koda K, Yamashita K, Kubo M, Kawakami F, Mitchell JB. Kagiya G, et al. Oncol Rep. 2015 Aug;34(2):1065-73. doi: 10.3892/or.2015.4020. Epub 2015 May 29. Oncol Rep. 2015. PMID: 26034980 Free PMC article. - CAIX-Mediated Control of LIN28/let-7 Axis Contributes to Metabolic Adaptation of Breast Cancer Cells to Hypoxia.
Gibadulinova A, Bullova P, Strnad H, Pohlodek K, Jurkovicova D, Takacova M, Pastorekova S, Svastova E. Gibadulinova A, et al. Int J Mol Sci. 2020 Jun 16;21(12):4299. doi: 10.3390/ijms21124299. Int J Mol Sci. 2020. PMID: 32560271 Free PMC article. - S-Nitrosylation in Tumor Microenvironment.
Sharma V, Fernando V, Letson J, Walia Y, Zheng X, Fackelman D, Furuta S. Sharma V, et al. Int J Mol Sci. 2021 Apr 27;22(9):4600. doi: 10.3390/ijms22094600. Int J Mol Sci. 2021. PMID: 33925645 Free PMC article. Review.
References
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. - PubMed
- Vaupel P, Mayer A, Hockel M. Tumor hypoxia and malignant progression. Methods Enzymol. 2004;381:335–354. - PubMed
- Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. The authors of this paper and ref. [4] performed ChIP-chip (chromatin immunoprecipitation and mRNA microarray) analysis to identify direct target genes of HIF-1 and HIF-2. The majority of hypoxia-induced genes contained HIF-1 binding sites and expression was dependent on HIF-1α expression. HIF-2α bound to many of the same genes, but knockdown of HIF-2α levels did not affect gene expression. Hypoxia-repressed gene expression was also HIF-1α-dependent but did not involve direct HIF-1α binding to target genes. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials