RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex - PubMed (original) (raw)
RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex
Caryn R Hale et al. Cell. 2009.
Abstract
Compelling evidence indicates that the CRISPR-Cas system protects prokaryotes from viruses and other potential genome invaders. This adaptive prokaryotic immune system arises from the clustered regularly interspaced short palindromic repeats (CRISPRs) found in prokaryotic genomes, which harbor short invader-derived sequences, and the CRISPR-associated (Cas) protein-coding genes. Here, we have identified a CRISPR-Cas effector complex that is comprised of small invader-targeting RNAs from the CRISPR loci (termed prokaryotic silencing (psi)RNAs) and the RAMP module (or Cmr) Cas proteins. The psiRNA-Cmr protein complexes cleave complementary target RNAs at a fixed distance from the 3' end of the integral psiRNAs. In Pyrococcus furiosus, psiRNAs occur in two size forms that share a common 5' sequence tag but have distinct 3' ends that direct cleavage of a given target RNA at two distinct sites. Our results indicate that prokaryotes possess a unique RNA silencing system that functions by homology-dependent cleavage of invader RNAs.
Figures
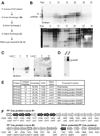
Figure 1. Identification of a ribonucleoprotein complex containing psiRNAs and Cas proteins
A) psiRNP purification scheme. Letters indicate the location of corresponding data within Figure 1. B) psiRNA (top panel) and total RNA (bottom panel) profiles across the initial Q-sepharose anion exchange fractions and an unfractionated sample (total). Northern analysis (top panel) was performed for P. furiosus psiRNA 7.01. The positions of the mature psiRNAs and 1X intermediate RNA (Hale et al., 2008) are indicated. The lower panel shows all RNAs detected by SYBR Gold staining. The peak fraction is indicated by an arrow in each panel. C) psiRNA (Northern analysis of psiRNA 7.01, left panel) and total RNA (SYBR staining, right panel) profiles across the S-sepharose cation exchange fractions and starting material (load). The peak fraction is indicated by an arrow in each panel. D) Native gel Northern analysis of the psiRNP. The peak S-sepharose fraction (arrow, C) was fractionated by native gel electrophoresis and analyzed by Northern blotting for psiRNA 7.01. RNA extracted from the same fraction was co-analyzed. The position of the psiRNP is indicated. E) Cas proteins identified by tandem mass spectrometry. The isolated psiRNP (D) was subject to in-gel trypsin digestion and tandem mass spectrometry. Sequence coverage and the number of unique peptides for Cas proteins identified with 99% confidence are shown. P. furiosus cas gene names are as given (Haft et al., 2005) and proposed functions are as predicted (Haft et al., 2005; Makarova et al., 2006). F) Genome organization of predicted P. furiosus cas genes. Operon organization and COG assignments were adapted from NCBI database. Core cas genes (cas) and Cas module-RAMP (cmr), Cas subtype Apern (csa) and Cas subtype Tneap (cst) genes are indicated. Proteins identified by mass spectrometry are indicated in black.
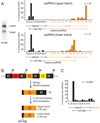
Figure 2. psiRNA species in the RNP contain a common 5’ sequence element and distinct 3’ termini
A) Sequence analysis of RNAs associated with the complex. RNA species present in the S-sepharose fraction (visualized by SYBR Gold staining) are shown in left panel. RNAs in the upper and lower bands were isolated, cloned, and sequenced. Graphs show the percentage of sequenced RNAs with 5’ ends located at specific positions within the repeat sequence (black), and with indicated numbers of guide sequence nucleotides downstream of the repeat sequence (orange). The average guide sequence is 37 nucleotides in P. furiosus. A consensus for each psiRNA species is diagrammed under each graph. The 8-nucleotide repeat sequence found at the 5’ end of the majority of the psiRNAs is indicated as the psi-tag. B) Model for biogenesis of the two psiRNA species in P. furiosus. CRISPR locus transcripts containing alternating repeat (R, black segments) and guide (G, colored segments) elements are cleaved at a specific site within the repeat by the Cas6 endoribonuclease (Carte et al., 2008), ultimately producing 1X intermediate RNAs that contain a full invader-targeting sequence flanked on both sides by segments of the repeat. The mature RNAs retain the 5’ end repeat sequence (psi-tag). Uncharacterized 3’ end processing of the 1X intermediate by endo- and/or exo-nucleases forms the two major mature psiRNAs: a 45-nucleotide species that contains the 8-nucleotide psi-tag and a full guide sequence, and a 39-nucleotide species that contains a shorter 31-nucleotide guide sequence. C) Deep sequencing of small RNAs from P. furiosus confirms the presence of the psi-tag. The 5’ ends of the sequenced psiRNAs are graphed as in A. The number of total clones analyzed (n) is indicated in the graphs of panels A and C.
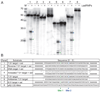
Figure 3. Specific cleavage of complementary target RNAs
The indicated 5’ end-labeled substrates were incubated in the presence (+) or absence (−) of the native psiRNPs (Figure 1C). Products were resolved by denaturing gel electrophoresis. The primary cleavage products are indicated by green and blue arrows (upper panel), and the corresponding sites of cleavage are indicated with green (site 1) and blue (site 2) vertical lines in the substrate sequences shown in the lower panel. The sizes of RNA markers (M) are indicated. “Target” substrates (panels 1, 3, 4, 5, 7) contain regions of perfect complementarity to the guide sequence of the indicated P. furiosus psiRNA. Grey bars demarcate the guide sequences in the lower panel. “+ ext” substrates (panels 1, 2, 3, 4, 7) contain 5’ and 3’ polyA extensions. In panel 4, a synthetic psiRNA (sequence shown in grey) was pre-annealed to the 7.01 target RNA + ext. Panel 2 shows a reverse target sequence substrate and panel 6 shows an antisense (AS) target substrate. Panel 3 shows a DNA substrate; all other substrates are RNA. Panel 8 shows unrelated RNA sR2.
Figure 4. Cleavages occurs 14 nucleotides from the 3’ ends of the psiRNAs
A)The indicated 5’ end-labeled (*) substrates were incubated in the presence (+) or absence (−) of the psiRNP (Figure 1C). The substrates were full-length 7.01 target RNA (F.L.), and the indicated truncations as diagramed in the lower panel. As in Figure 3, the locations of observed cleavages at sites 1 (green) and 2 (blue) are indicated on the full-length target and truncated RNAs (lower panel) and the corresponding cleavage products are indicated with green and blue arrows in the upper panels. Question mark in the lower panel indicates cleavage that could not be assessed. B) Model for cleavage at two sites directed by two psiRNAs. The 45-nucleotide psiRNA species guides cleavage at site 1 and the 39-nucleotide psiRNA guides cleavage at site 2 on each of the substrate RNAs as indicated. In both cases, cleavage occurs 14 nucleotides from the 3’ end of the psiRNA. Observed products are shown in green (site 1) and blue (site 2) and correspond to products in Figure 3 and Figure 4A.
Figure 5. Target RNA cleavage requires five Cmr proteins and a single psiRNA species
A) 5' end-labeled 7.01 target RNA was incubated in the absence of added psiRNAs or proteins (−), in the presence of synthetic psiRNAs (+) or purified recombinant P. furiosus Cmr proteins (R) or in the presence of purified native psiRNPs (N) as indicated. The synthetic psiRNAs were 45- and 39-nucleotide forms of psiRNA 7.01. The six added recombinant Cmr proteins were P. furiosus Cmr1-1, Cmr2, Cmr3, Cmr4, Cmr5 and Cmr6. Products were resolved by denaturing gel electrophoresis. The products corresponding to cleavage at site 1 and site 2 (see Figure 3) are indicated by green and blue arrows, respectively. The sizes of RNA markers (M) are indicated. B) The 7.01 target RNA (Target) was incubated with the synthetic 7.01 psiRNAs (both 45-and 39-nucleotide species) in the absence (+ psiRNAs) and presence of the purified recombinant P. furiosus Cmr proteins (all), and also with combinations of proteins lacking individual Cmr proteins as indicated (e.g. - Cmr6). C) Cleavage activity of the recombinant psiRNP (R) reconstituted with either the individual 7.01 psiRNA species (45- or 39-nt) or both. Cleavage by the native psiRNP (N) is included for comparison.
Figure 6. Model for the function of psiRNA-Cmr protein complexes in silencing molecular invaders
Based on the results of this study, a psiRNA with a conserved 5’ sequence element derived from the CRISPR repeat (psi-tag) and a region of invader-targeting sequence assembles with six Cas module-RAMP proteins (Cmr1-6). The assembled psiRNP interacts with an invader RNA (e.g., viral mRNA) through base pairing between the psiRNA and invader RNA, positioning the region of the RNA-RNA duplex 14 nucleotides from the 3’ end of the psiRNA in proximity to the active site (star) of the enzyme. In P. furiosus, there are two prominent size forms of each psiRNA with different 3’ ends that guide cleavage of viral mRNAs at two distinct sites. There are also two Cmr1 proteins in P. furiosus that are both found in purified preparations and likely function redundantly.
Comment in
- RNAi: prokaryotes get in on the act.
van der Oost J, Brouns SJ. van der Oost J, et al. Cell. 2009 Nov 25;139(5):863-5. doi: 10.1016/j.cell.2009.11.018. Cell. 2009. PMID: 19945373
Similar articles
- RNAi: prokaryotes get in on the act.
van der Oost J, Brouns SJ. van der Oost J, et al. Cell. 2009 Nov 25;139(5):863-5. doi: 10.1016/j.cell.2009.11.018. Cell. 2009. PMID: 19945373 - Structure of an RNA silencing complex of the CRISPR-Cas immune system.
Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Spilman M, et al. Mol Cell. 2013 Oct 10;52(1):146-52. doi: 10.1016/j.molcel.2013.09.008. Mol Cell. 2013. PMID: 24119404 Free PMC article. - Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus.
Hale C, Kleppe K, Terns RM, Terns MP. Hale C, et al. RNA. 2008 Dec;14(12):2572-9. doi: 10.1261/rna.1246808. Epub 2008 Oct 29. RNA. 2008. PMID: 18971321 Free PMC article. - The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus.
Terns RM, Terns MP. Terns RM, et al. Biochem Soc Trans. 2013 Dec;41(6):1416-21. doi: 10.1042/BST20130056. Biochem Soc Trans. 2013. PMID: 24256230 Free PMC article. Review. - CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation.
Bhaya D, Davison M, Barrangou R. Bhaya D, et al. Annu Rev Genet. 2011;45:273-97. doi: 10.1146/annurev-genet-110410-132430. Annu Rev Genet. 2011. PMID: 22060043 Review.
Cited by
- Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex.
Shao Y, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. Shao Y, et al. Structure. 2013 Mar 5;21(3):376-84. doi: 10.1016/j.str.2013.01.002. Epub 2013 Feb 7. Structure. 2013. PMID: 23395183 Free PMC article. - The structural biology of CRISPR-Cas systems.
Jiang F, Doudna JA. Jiang F, et al. Curr Opin Struct Biol. 2015 Feb;30:100-111. doi: 10.1016/j.sbi.2015.02.002. Epub 2015 Feb 24. Curr Opin Struct Biol. 2015. PMID: 25723899 Free PMC article. Review. - Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins.
Jia N, Patel DJ. Jia N, et al. Nat Rev Mol Cell Biol. 2021 Aug;22(8):563-579. doi: 10.1038/s41580-021-00371-9. Epub 2021 Jun 4. Nat Rev Mol Cell Biol. 2021. PMID: 34089013 Review. - Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management.
Allemailem KS, Alsahli MA, Almatroudi A, Alrumaihi F, Alkhaleefah FK, Rahmani AH, Khan AA. Allemailem KS, et al. Cancer Commun (Lond). 2022 Dec;42(12):1257-1287. doi: 10.1002/cac2.12366. Epub 2022 Oct 9. Cancer Commun (Lond). 2022. PMID: 36209487 Free PMC article. Review. - Repeat Size Determination by Two Molecular Rulers in the Type I-E CRISPR Array.
Goren MG, Doron S, Globus R, Amitai G, Sorek R, Qimron U. Goren MG, et al. Cell Rep. 2016 Sep 13;16(11):2811-2818. doi: 10.1016/j.celrep.2016.08.043. Cell Rep. 2016. PMID: 27626652 Free PMC article.
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. A Novel Family of Sequence-specific Endoribonucleases Associated with the Clustered Regularly Interspaced Short Palindromic Repeats. J Biol Chem. 2008;283:20361–20371. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054682-11/GM/NIGMS NIH HHS/United States
- R01 GM062516/GM/NIGMS NIH HHS/United States
- R01 GM054682/GM/NIGMS NIH HHS/United States
- R01GM54682/GM/NIGMS NIH HHS/United States
- R01GM062516/GM/NIGMS NIH HHS/United States
- R01 GM062516-08/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases