Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis - PubMed (original) (raw)
Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis
Binglian Zheng et al. Genes Dev. 2009.
Abstract
Intergenic transcription by RNA Polymerase II (Pol II) is widespread in plant and animal genomes, but the functions of intergenic transcription or the resulting noncoding transcripts are poorly understood. Here, we show that Arabidopsis Pol II is indispensable for endogenous siRNA-mediated transcriptional gene silencing (TGS) at intergenic low-copy-number loci, despite the presence of two other polymerases-Pol IV and Pol V-that specialize in TGS through siRNAs. We show that Pol II produces noncoding scaffold transcripts that originate outside of heterochromatic, siRNA-generating loci. Through these transcripts and physical interactions with the siRNA effector protein ARGONAUTE4 (AGO4), Pol II recruits AGO4/siRNAs to homologous loci to result in TGS. Meanwhile, Pol II transcription also recruits Pol IV and Pol V to different locations at heterochromatic loci to promote siRNA biogenesis and siRNA-mediated TGS, respectively. This study establishes that intergenic transcription by Pol II is required for siRNA-mediated TGS, and reveals an intricate collaboration and division of labor among the three polymerases in gene silencing.
Figures
Figure 1.
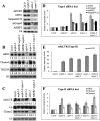
Isolation of an nrpb2 mutant. (A) Three-week-old seedlings of various genotypes as indicated. (NRPB2 nrpb2-3) An nrpb2-3 mutant carrying an NRPB2 transgene. Note that the photos of the mutant plants were taken at a higher magnification than those of the wild-type and the rescued line. (B) Partial amino acid sequence of RPB2 in various species. The conserved glycine, which was mutated to glutamic acid in nrpb2-3, is marked by a rectangle and an asterisk. The other rectangles show GW/WG motifs in RPB2 proteins. (At) Arabidopsis thaliana; (Sp) Schizosaccharomyces pombe; (Hs) Homo sapiens; (Mm) Mus musculus; (Dm) Drosophila melanogaster; (Ce) Caenorhabditis elegans. (C) Western blotting to determine the levels of NRPB2 and NRPB1 in wild type (Col-0) and nrpb2-3. HEN1 and Hsp70 served as loading controls.
Figure 2.
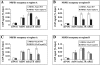
siRNA accumulation and TGS at heterochromatic loci in various genotypes. (A–C) siRNA accumulation in various genotypes. (Col-0) Wild type; (nrpb2-3) a Pol II mutant; (sde4-3) a Pol IV mutant; (nrpe1-1) a Pol V mutant; (NRPB2 nrpb2-3) the nrpb2-3 mutant complemented with NRPB2 genomic DNA; two independent transgenic lines were included in B. The numbers below the gel images indicate the relative levels of an siRNA among the different genotypes. The U6 blots served as loading controls for the small RNA blots above. (D–F) Real-time RT–PCR analysis of the expression of siRNA loci in various genotypes. Transcripts from the siRNA-generating region (region A in Fig. 3A) were detected. Transcript levels were normalized to those of UBQ5 and the mutants were compared with wild type. Standard deviations were calculated from three technical replicates. The results shown were reproduced with three biological replicates.
Figure 3.
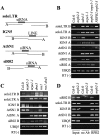
Detection of noncoding transcripts at siRNA loci. (A) Diagrams of soloLTR, IGN5, AtSN1, and siR02 genomic regions. The soloLTR, IGN5, and AtSN1 regions are based on analysis of transcription units by Wierzbicki et al. (2008). (B,C) RT–PCR analysis of noncoding transcripts at siRNA loci in various genotypes. (D) RIP with anti-RPB2 antibodies to test the association of Pol II with noncoding transcripts at siRNA loci. In B–D, UBQ5 served as a loading control. The RT (−) control PCR was performed with UBQ5 primers. The results shown were reproduced at least three times.
Figure 4.
Pol II is present at siRNA loci. (A,B) Real-time PCR of DNA that copurified with Pol II in ChIP performed in various genotypes with an anti-RPB1 antibody. The eIF4A1 gene served as a positive control. (C,D) The “No Ab” (no antibody) immunoprecipitate served as negative controls. The ChIP signal was quantified as the percentage of total input DNA. Two biological replicates were performed and identical results were obtained. Standard deviations were calculated from three technical repeats.
Figure 5.
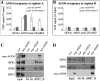
Recruitment of AGO4 by Pol II. (A,B) AGO4 occupancy at regions A and B of siRNA loci. ChIP was performed using an anti-myc antibody and DNA that copurified with myc-AGO4 was quantified by real-time PCR. Region A, but not B, was enriched in the myc-AGO4 immunoprecipitate, as compared with the eIF4A1 negative control. (WT) Wild type. (C,D) Co-IP to test the association between AGO4 and Pol II in vivo with anti-RPB1 and anti-myc antibodies, respectively. (L_er_) Wild type; a negative control for myc-AGO4. Proteins from one-sixth of “No Ab” (no antibody) immunoprecipitate and one-sixth of immunoprecipitate were analyzed by Western blotting. Proteins from 1/100 of input were included for anti-RPB1, anti-RPB2, and anti-HEN1 blots, while proteins from 1/1000 of input were used for the anti-myc blot.
Figure 6.
Pol V and Pol IV occupancy at regions A and B of siRNA loci. ChIP was performed against NRPE1-Flag (A,B) or NRPD1-Flag (C,D), and the coprecipitated DNA was quantified by real-time PCR and expressed as the percentage of input DNA. eIF4A1 served as a negative control. Standard deviations were calculated from three technical repeats. The results shown were reproduced in two biological replicates. (WT) Wild type.
Figure 7.
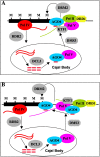
Models for the roles of the three polymerases in TGS at type II loci in Arabidopsis. In both models, no modifications to the prevailing views on the roles of Pol IV, RDR2, DCL3, and AGO4 are made except that we showed that Pol IV is associated with the chromatin at siRNA loci. The two models differ in the roles of Pol II and Pol V in TGS. (A) Pol II generates scaffold transcripts that originate in region B and that traverse region A. The Pol II transcripts or Pol II transcription itself recruits Pol V, which is in complex with AGO4 during siRNA biogenesis in the Cajal body, to region B. Pol V delivers AGO4 to Pol II and AGO4/siRNA RISCs localize to region A due to the sequence complementarity between siRNAs, which are only derived from region A, to Pol II transcripts. (B) Pol II generates scaffold transcripts that correspond only to region B. These transcripts recruit Pol V to region B and Pol V makes new scaffold transcripts that traverse region A. These transcripts help anchor AGO4/siRNAs to region A. In both models, AGO4/siRNA RISCs then recruit, directly or indirectly, the DNA methyltransferase DRM2 or histone methyltransferases to result in epigenetic modifications and silencing. KTF1 may bind to Pol II transcripts to facilitate the recruitment of Pol V by Pol II transcripts. DRD1 is required for the production of Pol II transcripts. DMS3 acts downstream from Pol II transcripts to facilitate the recruitment of AGO4. (M) DNA methylation.
Similar articles
- Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana.
Sasaki T, Lee TF, Liao WW, Naumann U, Liao JL, Eun C, Huang YY, Fu JL, Chen PY, Meyers BC, Matzke AJ, Matzke M. Sasaki T, et al. Plant J. 2014 Jul;79(1):127-38. doi: 10.1111/tpj.12545. Epub 2014 Jun 13. Plant J. 2014. PMID: 24798377 - NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes.
Wierzbicki AT, Haag JR, Pikaard CS. Wierzbicki AT, et al. Cell. 2008 Nov 14;135(4):635-48. doi: 10.1016/j.cell.2008.09.035. Cell. 2008. PMID: 19013275 Free PMC article. - Finding the right template: RNA Pol IV, a plant-specific RNA polymerase.
Vaughn MW, Martienssen RA. Vaughn MW, et al. Mol Cell. 2005 Mar 18;17(6):754-6. doi: 10.1016/j.molcel.2005.03.003. Mol Cell. 2005. PMID: 15780931 Review. - The role of long non-coding RNA in transcriptional gene silencing.
Wierzbicki AT. Wierzbicki AT. Curr Opin Plant Biol. 2012 Nov;15(5):517-22. doi: 10.1016/j.pbi.2012.08.008. Epub 2012 Sep 6. Curr Opin Plant Biol. 2012. PMID: 22960034 Review.
Cited by
- A conserved Pol II elongator SPT6L mediates Pol V transcription to regulate RNA-directed DNA methylation in Arabidopsis.
Liu Y, Shu J, Zhang Z, Ding N, Liu J, Liu J, Cui Y, Wang C, Chen C. Liu Y, et al. Nat Commun. 2024 May 25;15(1):4460. doi: 10.1038/s41467-024-48940-8. Nat Commun. 2024. PMID: 38796517 Free PMC article. - Recent Advances in Studies of Genomic DNA Methylation and Its Involvement in Regulating Drought Stress Response in Crops.
Fan Y, Sun C, Yan K, Li P, Hein I, Gilroy EM, Kear P, Bi Z, Yao P, Liu Z, Liu Y, Bai J. Fan Y, et al. Plants (Basel). 2024 May 17;13(10):1400. doi: 10.3390/plants13101400. Plants (Basel). 2024. PMID: 38794470 Free PMC article. Review. - Mapping nucleosome-resolution chromatin organization and enhancer-promoter loops in plants using Micro-C-XL.
Sun L, Zhou J, Xu X, Liu Y, Ma N, Liu Y, Nie W, Zou L, Deng XW, He H. Sun L, et al. Nat Commun. 2024 Jan 2;15(1):35. doi: 10.1038/s41467-023-44347-z. Nat Commun. 2024. PMID: 38167349 Free PMC article. - Comparative analysis of nascent RNA sequencing methods and their applications in studies of cotranscriptional splicing dynamics.
Liu M, Zhu J, Huang H, Chen Y, Dong Z. Liu M, et al. Plant Cell. 2023 Nov 30;35(12):4304-4324. doi: 10.1093/plcell/koad237. Plant Cell. 2023. PMID: 37708036 Free PMC article. - Plant polymerase IV sensitizes chromatin through histone modifications to preclude spread of silencing into protein-coding domains.
Hari Sundar G V, Swetha C, Basu D, Pachamuthu K, Raju S, Chakraborty T, Mosher RA, Shivaprasad PV. Hari Sundar G V, et al. Genome Res. 2023 May;33(5):715-728. doi: 10.1101/gr.277353.122. Epub 2023 Jun 5. Genome Res. 2023. PMID: 37277199 Free PMC article.
References
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. - PubMed
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases