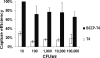Oriented immobilization of bacteriophages for biosensor applications - PubMed (original) (raw)
Oriented immobilization of bacteriophages for biosensor applications
M Tolba et al. Appl Environ Microbiol. 2010 Jan.
Abstract
A method was developed for oriented immobilization of bacteriophage T4 through introduction of specific binding ligands into the phage head using a phage display technique. Fusion of the biotin carboxyl carrier protein gene (bccp) or the cellulose binding module gene (cbm) with the small outer capsid protein gene (soc) of T4 resulted in expression of the respective ligand on the phage head. Recombinant bacteriophages were characterized in terms of infectivity. It was shown that both recombinant phages retain their lytic activity and host range. However, phage head modification resulted in a decreased burst size and an increased latent period. The efficiency of bacteriophage immobilization with streptavidin-coated magnetic beads and cellulose-based materials was investigated. It was shown that recombinant bacteriophages form specific and strong bonds with their respective solid support and are able to specifically capture and infect the host bacterium. Thus, the use of immobilized BCCP-T4 bacteriophage for an Escherichia coli B assay using a phage multiplication approach and real-time PCR allowed detection of as few as 800 cells within 2 h.
Figures
FIG. 1.
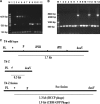
Detection of soc insertion into the T4 genome by use of PCR. The primers at the 3′ end of gene e and the 3′ end of gene denV were used in the amplification reactions. (A) Lanes 3 to 7, 1.3-kb PCR products indicating the insertion of soc-bccp; lanes 8 to 12, fragment obtained using T4-Z phage; lane M, molecular size standard. (B) Lanes 7, 11, and 14, 2-kb PCR product indicating the presence of CBM-GFP fusions; lanes 1 to 6, 8 to 10, and 12 and 13, 0.5-kb DNA fragment obtained using T4-Z phage; lane M, molecular size standard. (C) Schematic presentation of the constructs used in the study.
FIG. 2.
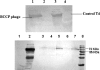
Expression of the affinity tags on the phage head as detected by Western blotting. (A) Verification of biotinylation. Lanes 1 and 3, BCCP-T4 phage propagated in E. coli AVB-100 overexpressing BirA; lanes 2 and 4, T4 phage. (B) Verification of expression of green fluorescent protein on CBM-T4 phage by use of anti-GFP antibodies. Lane 1, E. coli B; lane 2, E. coli cells containing the cbm-gfp plasmid; lanes 3 to 5, and 7, CBM-T4 phage; lane 6, T4-Z phage (control); lane 8, molecular size standard.
FIG. 3.
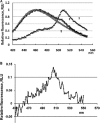
(A) Normalized fluorescent spectra of E. coli expressing CBM-GFP (1), CBM-T4 phage (2), and T4 phage (3) (excitation at 395 nm). (B) Differential fluorescence spectra for CBM-T4 and T4 bacteriophages (excitation at 395 nm).
FIG. 4.
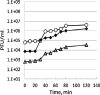
One-step growth curves for bacteriophages T4 (⧫), BCCP-T4 (○), and CBM-T4 (▵).
FIG. 5.
Scanning electron micrographs of BCCP-T4 phage immobilized on the streptavidin-coated Au surface. Arrows point to the phage particles oriented with tails up.
FIG. 6.
Percentages of E. coli B cells captured by the immobilized phage. A portion (20 μl) of the constructed biosorbent (109 beads/ml) was used to capture the cells in 1 ml of LB broth. The time of contact was −10 min. Error bars show standard deviations.
FIG. 7.
Real-time PCR detection of progeny phage produced in E. coli. The number of cells refers to the sample volume (6 μl). CF RFU, curve fit relative fluorescence units.
Similar articles
- Bacteriophage-based biosorbents coupled with bioluminescent ATP assay for rapid concentration and detection of Escherichia coli.
Minikh O, Tolba M, Brovko LY, Griffiths MW. Minikh O, et al. J Microbiol Methods. 2010 Aug;82(2):177-83. doi: 10.1016/j.mimet.2010.05.013. Epub 2010 Jun 1. J Microbiol Methods. 2010. PMID: 20561957 - Development of a novel bacteriophage based biomagnetic separation method as an aid for sensitive detection of viable Escherichia coli.
Wang Z, Wang D, Chen J, Sela DA, Nugen SR. Wang Z, et al. Analyst. 2016 Feb 7;141(3):1009-16. doi: 10.1039/c5an01769f. Epub 2015 Dec 22. Analyst. 2016. PMID: 26689710 - T4 bacteriophage as a phage display platform.
Gamkrelidze M, Dąbrowska K. Gamkrelidze M, et al. Arch Microbiol. 2014 Jul;196(7):473-9. doi: 10.1007/s00203-014-0989-8. Epub 2014 May 15. Arch Microbiol. 2014. PMID: 24828789 Free PMC article. Review. - Xenogeneic Regulation of the Bacterial Transcription Machinery.
Tabib-Salazar A, Mulvenna N, Severinov K, Matthews SJ, Wigneshweraraj S. Tabib-Salazar A, et al. J Mol Biol. 2019 Sep 20;431(20):4078-4092. doi: 10.1016/j.jmb.2019.02.008. Epub 2019 Feb 15. J Mol Biol. 2019. PMID: 30776429 Review.
Cited by
- Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth.
Hyman P. Hyman P. Pharmaceuticals (Basel). 2019 Mar 11;12(1):35. doi: 10.3390/ph12010035. Pharmaceuticals (Basel). 2019. PMID: 30862020 Free PMC article. Review. - Bacteriophage Capsid Modification by Genetic and Chemical Methods.
Carmody CM, Goddard JM, Nugen SR. Carmody CM, et al. Bioconjug Chem. 2021 Mar 17;32(3):466-481. doi: 10.1021/acs.bioconjchem.1c00018. Epub 2021 Mar 4. Bioconjug Chem. 2021. PMID: 33661607 Free PMC article. Review. - Formulations for Bacteriophage Therapy and the Potential Uses of Immobilization.
Rosner D, Clark J. Rosner D, et al. Pharmaceuticals (Basel). 2021 Apr 13;14(4):359. doi: 10.3390/ph14040359. Pharmaceuticals (Basel). 2021. PMID: 33924739 Free PMC article. Review. - In Vivo Capsid Engineering of Bacteriophages for Oriented Surface Conjugation.
Hufziger KA, Farquharson EL, Werner BG, Chen Q, Goddard JM, Nugen SR. Hufziger KA, et al. ACS Appl Bio Mater. 2022 Nov 21;5(11):5104-5112. doi: 10.1021/acsabm.2c00428. Epub 2022 Oct 20. ACS Appl Bio Mater. 2022. PMID: 36264000 Free PMC article. - Recent advances in bacteriophage based biosensors for food-borne pathogen detection.
Singh A, Poshtiban S, Evoy S. Singh A, et al. Sensors (Basel). 2013 Jan 30;13(2):1763-86. doi: 10.3390/s130201763. Sensors (Basel). 2013. PMID: 23364199 Free PMC article. Review.
References
- Bennett, A. R., G. G. C. Davis, S. Vlahodimous, J. C. Banks, and P. R. Bell. 1997. Use of bacteriophages-based system for separation and concentration of Salmonella. J. Appl. Microbiol. 83:259-265. - PubMed
- Black, L. W., M. K. Showe, and A. C. Steven. 1994. Morphogenesis of the T4 head, p. 218-258. In J. Karam (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, DC.
- Blasco, R., M. J. Murphy, M. F. Sanders, and D. J. Squirrell. 1998. Specific assays for bacteria using phage mediated release of adenylate kinase. J. Appl. Microbiol. 84:661-666. - PubMed
- Boraston, A. B., A. L. Creagh, M. M. Alam, J. M. Kormos, P. Tomme, C. A. Haynes, and D. G. Kilburn. 2001. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40:6240-6247. - PubMed
- Camara, F. P., and D. F. Dealmeido. 1991. Studies on the growth of the phage T4 in cell division mutants DV1211 and AD651 of Escherichia coli at normal and high temperature. Rev. Bras. Genet. 14:233-238.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources