Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3' UTR - PubMed (original) (raw)
Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3' UTR
Niankun Liu et al. Genes Dev. 2009.
Abstract
Vasa (Vas) is a DEAD-box RNA-binding protein required in Drosophila at several steps of oogenesis and for primordial germ cell (PGC) specification. Vas associates with eukaryotic initiation factor 5B (eIF5B), and this interaction has been implicated in translational activation of gurken mRNA in the oocyte. Vas is expressed in all ovarian germline cells, and aspects of the vas-null phenotype suggest a function in regulating the balance between germline stem cells (GSCs) and their fate-restricted descendants. We used a biochemical approach to recover Vas-associated mRNAs and obtained mei-P26, whose product represses microRNA activity and promotes GSC differentiation. We found that vas and mei-P26 mutants interact, and that mei-P26 translation is substantially reduced in vas mutant cells. In vitro, Vas protein bound specifically to a (U)-rich motif in the mei-P26 3' untranslated region (UTR), and Vas-dependent regulation of GFP-mei-P26 transgenes in vivo was dependent on the same (U)-rich 3' UTR domain. The ability of Vas to activate mei-P26 expression in vivo was abrogated by a mutation that greatly reduces its interaction with eIF5B. Taken together, our data support the conclusion that Vas promotes germ cell differentiation by directly activating mei-P26 translation in early-stage committed cells.
Figures
Figure 1.
Purification of Vas–mRNA complexes. (A) Aliquots (1/100) of eluates from oligo(dT) chromatography were resolved on a 0.75% TAE agarose gel, blotted, and treated with anti-Vas antibody. (Lanes 1–3) First, second, and third eluates from uncross-linked controls. (Lanes 4–6) First, second, and third eluates from cross-linked samples. (Lane 7) Cross-linked lysate before oligo(dT) chromatography, a positive control. (B) Purification of Vas–mRNA complexes by tandem Vas immunoprecipitation, analyzed by immunoblot as in A. (Lane 1) Cross-linked sample before immunoprecipitation, a positive control. (Lanes 2,3) Aliquots of material recovered from the first immunoprecipitation, using rabbit IgG (control, lane 2) and α-Vas (lane 3). (Lanes 4,5) Aliquots of material recovered from the second immunoprecipitation using rabbit IgG (control, lane 4) and α-Vas (lane 5). The arrows indicate the purified Vas–mRNA complexes. (C) Synthesis of double-strand cDNA. Lane 1 is a positive control. Lanes 2–4 are the first-strand cDNA synthesis, and represent buffer control (lane 2), uncross-linked control (lane 3), and cross-linked sample (lane 4). Lanes 5–7 are the second-strand cDNA synthesis, and represent buffer control (lane 5), uncross-linked control (lane 6), and cross-linked sample (lane 7).
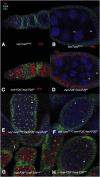
Figure 2.
mei-P26 and vas mutations genetically interact. Nuclei were visualized by DAPI staining (blue), germline cells were labeled with α-Vas, and fusomes were labeled with mAb 1B1. (A,B) vas1/vasPH165 egg chambers are similar to wild-type, composed of 15 nurse cells (nc) and an oocyte (oo), surrounded by a layer of follicle cells. (C,D) mei-P261/mei-P26fs1 ovaries, a relatively mild allelic combination, are mostly wild-type in appearance. Occasional egg chambers (<5%) still differentiate nurse cells and oocytes, but have fewer or more than 15 nurse cells, or are incorrectly patterned (data not shown). (_E_,_F_) _vas1_/_vasPH165_; _mei-P261_/_mei-P26fs1_ ovaries have a much more severe phenotype, with most (>50%) egg chambers appearing tumorous and failing to differentiate nurse cells and oocytes. The germline cells of these ovaries show either punctate spectrosomes (asterisks) or branched fusomes (solid triangles), indicating they are cystocyte-derived. (G,H) ovaries from mei-P26fs1/mei-P26mfs1, a strong allelic combination, resemble vas1/vasPH165; mei-P261/mei-P26fs1 ovaries.
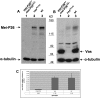
Figure 3.
Mei-P26 protein level is reduced in vas mutant ovaries. (A) Immunoblot of ovarian extracts from mei-P26fs1/mei-P26mfs1, vasPH165, and wild-type (OreR) females using anti-Mei-P26, which was preadsorbed with fixed mei-P26fs1/mei-P26mfs1 ovaries. The same blot was probed with anti-α-tubulin as a loading control. (Lane 1) mei-P26fs1/mei-P26mfs1 ovarian extract serves as a negative control. (Lane 2) Homozygous vasPH165 ovarian extract. (Lane 3) Wild-type ovarian extract. The Mei-P26 level is reduced in lane 2 as compared with lane 3 (arrow). (B) Immunoblot using anti-Vas on extracts prepared from mei-P26fs1/mei-P26mfs1 ovaries (lane 1), homozygous vasPH165 ovaries (lane 2), and wild-type ovaries (lane 3). The Vas level (arrow) is unaffected by mei-P26 mutations. Molecular weight markers are indicated in kilodaltons. (C) Real-time PCR data comparing mei-P26 RNA levels in mei-P26fs1/mei-P26mfs1 (mei-P26), vasPH165 (vas), and wild-type (wt) ovaries, showing that mei-P26 RNA levels are unaffected by vas mutation. The data presented are averaged from four independent experiments (12 reactions) for each genotype, and the error bars indicate standard deviations.
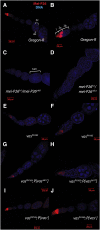
Figure 4.
Vas activity affects Mei-P26 protein accumulation. (A,B) In wild-type ovaries, Mei-P26 protein is present in stem cells, and its expression reaches a peak in 16-cell cysts. It is essentially absent in subsequent stages of oogenesis. In A, an egg chamber, which in normal development includes 15 polyploid nurse cells (nc) and an oocyte (oo), is labeled. In B, the different developmental stages of the germarium are labeled. Region 1 (1) contains the stem cells and the mitotic region in which the cystocytes divide to form two-, four-, eight-, and 16-cell cysts; region 2a (2a) contains early 16-cell cysts, which mature in region 2b (2b), becoming associated with a monolayer of precursor follicle cells. Region 2b is also the region in which nurse cells and oocytes differentiate (Spradling et al. 1997). Mei-P26 is expressed in region 1, peaking in early 16-cell cysts. (C,D) Little Mei-P26 signal is apparent in mei-P26fs1/_mei-P26mfs1_ovaries. These egg chambers are tumorous (tum), failing to differentiate nurse cells and oocytes. (E,F) Accumulation of Mei-P26 protein throughout its expression domain is substantially reduced in homozygous vasPH165 ovaries. (G,H) Accumulation of Mei-P26 protein is similarly reduced in vasPH165; P{vasΔ617} ovaries. (I,J) Mei-P26 protein expression in vasPH165 ovaries is rescued to wild-type levels by expression of a wild-type vas+ transgene.
Figure 5.
RNA-binding assays reveal a (U)-rich motif that is recognized specifically by full-length GST-Vas. (A) Gel retardation assays to investigate the RNA-binding properties of Vas in respect to mei-P26 mRNA. GST (1.5 μg) or GST-Vas (+, 2 μg; or ++, 4 μg) was incubated with 30 ng of 32P-labeled RNA probes as follows: (Lanes 1–3) Full-length mei-P26 5′ UTR RNA; (lanes 4–6) Full-length mei-P26 3′ UTR RNA (antisense strand). (Lanes 7–9,16–19) Full-length mei-P26 3′ UTR RNA (sense strand). (Lanes 10–12) mei-P26 3′ UTR RNA deleted for the (U)-rich element (nucleotides 565–574). (Lanes 13–15) mei-P26 3′ UTR RNA deleted for a larger segment containing the (U)-rich element (nucleotides 551–580). Unlabeled poly(U) was added to the samples loaded in lanes 18 and 19 (0.1 μg and 1.0 μg, respectively). RNAs were separated on a 0.7% agarose gel. Species with retarded electrophoretic mobility (GS) were observed only when GST-Vas and full-length mei-P26 3′ UTR were present, and poly(U) competitor RNA was absent. In those reactions, the molar ratio of GST:Vas to mei-P26 3′ UTR RNA was 325:1. (B) Cross-linking assays to test the ability of Vas to bind nucleotides 551–580 of the mei-P26 3′ UTR in the presence of various competitor RNAs. Lanes 1 and 2 are negative controls (no added protein and GST added, respectively). (Lanes 3–14) GST-Vas (4 μg) and 32P-labeled mei-P26 3′ UTR nucleotides 551–580 (20 ng) were UV-cross-linked in the presence of competitors as follows: (Lanes 3–6) mei-P26 3′ UTR nucleotides 551–580. (Lanes 7–10) Poly(U). (Lanes 11–14)Poly(A). Competitor RNAs were added at 0 μg (labeled 0), 0.01 μg (+), 0.1 μg (++), and 1 μg (+++). The position at which samples were loaded onto the gel is labeled “top” and the position at which the major cross-linked species migrates is labeled “XL.” (C) Cross-linking assays to compare the relative affinity of full-length GST-Vas to nucleotides 551–580 of the mei-P26 3′ UTR to its affinity to neighboring 30-nt fragments of the mei-P26 3′ UTR. GST (1.5 μg) or GST-Vas (+, 2 μg; or ++, 4 μg) was incubated with the following 32P-labeled (30-ng) RNA probes as follows: (Lanes 1–3) mei-P26 3′ UTR nucleotides 521–550. (Lanes 4–6) mei-P26 3′ UTR nucleotides 551–580. (Lanes 7–9) mei-P26 3′ UTR nucleotides 581–610. (Lanes 10–12) mei-P26 3′ UTR nucleotides 611–640. The position at which samples were loaded onto the gel is labeled “top” and the position at which the major cross-linked species migrates is labeled “XL.” The intensities of the major cross-linked species were measured with ImageQuant software, and are presented as a percentage of that measured in lane 6. In this experiment, when 4 μg of GST-Vas were present, the molar ratio of protein to RNA was 12:1. (D) Cross-linking assays as in C but using 4 μg of GST-VCP instead of the full-length GST-Vas, and using 20 ng of the RNA probes. Unlike GST-Vas, GST-VCP does not preferentially bind the 551–580 segment. In this experiment, when 4 μg of GST-VCP were present, the molar ratio of protein to RNA was 25:1. All of the transcripts used in this figure derive from a mei-P26 cDNA that contains 10 consecutive U residues in its 3′ UTR (nucleotides 565–574), but that differs from the corresponding region of the consensus genome sequence, which has only nine consecutive T residues (Celniker et al. 2002).
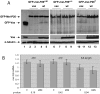
Figure 6.
The Vas-binding site in the mei-P26 3′ UTR affects GFP-Mei-P26 expression in vivo. (A) Immunoblot of ovarian extracts from various genotypes. α-GFP was used to detect GFP-Mei-P26 expressed from transgenes as indicated. (vas) Transgene is expressed in a vasPH165 genetic background; (wt) transgene is expressed is a wild-type genetic background. α-Vas was used to confirm the genotypes, and anti-α-tubulin was used as a loading control. Quantitative data from four independent experiments are presented in the Results. (Lane 1) P[_GFP-vas_] serves as positive control for α-GFP. (Lanes 2,3) vasPH165; P[_GFP-mei-P26Δ30_]. (Lanes 4,5) +; P[_GFP-mei-P26Δ30_]. The level of GFP-Mei-P26 is equivalent in wild-type and vas mutant backgrounds. (Lanes 6,7) vasPH165; P[_GFP-mei-P26Δ10_]. (Lanes 8,9) +; P[_GFP-mei-P26Δ10_]. The level of GFP-Mei-P26 is equivalent in wild-type and vas mutant backgrounds. (Lanes 10,11) vasPH165; P[_GFP-mei-P26_]. (Lanes 12,13) +; P[_GFP-mei-P26_]. The level of GFP-Mei-P26 is reduced in the vas mutant background as compared with wild-type, indicating that the Vas-binding site is essential for Vas-mediated activation of expression. (B) Real-time PCR data comparing mei-P26 RNA levels from the same transgenic constructs in wild-type and in vasPH165 genetic backgrounds. mRNA levels from all three constructs are slightly reduced in vasPH165 extracts, to 86.6% for full-length, 96.5% for GFP-mei-P26Δ10, and 92.8% for GFP-mei-P26Δ30. The _P_-values for these differences are given for each construct.
Similar articles
- Multiple Functions of the DEAD-Box Helicase Vasa in Drosophila Oogenesis.
Dehghani M, Lasko P. Dehghani M, et al. Results Probl Cell Differ. 2017;63:127-147. doi: 10.1007/978-3-319-60855-6_6. Results Probl Cell Differ. 2017. PMID: 28779316 Review. - WD40 protein Wuho controls germline homeostasis via TRIM-NHL tumor suppressor Mei-p26 in Drosophila.
Rastegari E, Kajal K, Tan BS, Huang F, Chen RH, Hsieh TS, Hsu HJ. Rastegari E, et al. Development. 2020 Jan 15;147(2):dev182063. doi: 10.1242/dev.182063. Development. 2020. PMID: 31941704 Free PMC article. - Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling.
Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M. Li Y, et al. Development. 2012 May;139(9):1547-56. doi: 10.1242/dev.077412. Epub 2012 Mar 21. Development. 2012. PMID: 22438571 Free PMC article. - Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila.
Chen D, Wu C, Zhao S, Geng Q, Gao Y, Li X, Zhang Y, Wang Z. Chen D, et al. PLoS Genet. 2014 Nov 20;10(11):e1004797. doi: 10.1371/journal.pgen.1004797. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25412508 Free PMC article. - Germline specification: small things have a big role.
Jin Z, Xie T. Jin Z, et al. Curr Biol. 2006 Nov 21;16(22):R966-7. doi: 10.1016/j.cub.2006.10.018. Curr Biol. 2006. PMID: 17113380 Review.
Cited by
- Translational control in cellular and developmental processes.
Kong J, Lasko P. Kong J, et al. Nat Rev Genet. 2012 Jun;13(6):383-94. doi: 10.1038/nrg3184. Nat Rev Genet. 2012. PMID: 22568971 Review. - A regulatory network of Drosophila germline stem cell self-renewal.
Yan D, Neumüller RA, Buckner M, Ayers K, Li H, Hu Y, Yang-Zhou D, Pan L, Wang X, Kelley C, Vinayagam A, Binari R, Randklev S, Perkins LA, Xie T, Cooley L, Perrimon N. Yan D, et al. Dev Cell. 2014 Feb 24;28(4):459-73. doi: 10.1016/j.devcel.2014.01.020. Dev Cell. 2014. PMID: 24576427 Free PMC article. - C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs.
Beshore EL, McEwen TJ, Jud MC, Marshall JK, Schisa JA, Bennett KL. Beshore EL, et al. Dev Biol. 2011 Feb 15;350(2):370-81. doi: 10.1016/j.ydbio.2010.12.005. Epub 2010 Dec 10. Dev Biol. 2011. PMID: 21146518 Free PMC article. - Mei-P26 mediates tissue-specific responses to the Brat tumor suppressor and the dMyc proto-oncogene in Drosophila.
Ferreira A, Boulan L, Perez L, Milán M. Ferreira A, et al. Genetics. 2014 Sep;198(1):249-58. doi: 10.1534/genetics.114.167502. Epub 2014 Jul 1. Genetics. 2014. PMID: 24990993 Free PMC article. - Germ plasm localisation of the HELICc of Vasa in Drosophila: analysis of domain sufficiency and amino acids critical for localisation.
Wang SC, Hsu HJ, Lin GW, Wang TF, Chang CC, Lin MD. Wang SC, et al. Sci Rep. 2015 Sep 30;5:14703. doi: 10.1038/srep14703. Sci Rep. 2015. PMID: 26419889 Free PMC article.
References
- Abdu U, Brodsky M, Schüpbach T. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of gurken through Chk2/Mnk. Curr Biol. 2002;12:1645–1651. - PubMed
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuléen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. - PubMed
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jäckle H, Lasko P. Vasa mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. - PubMed
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. - DOI - PMC - PubMed
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous