DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation - PubMed (original) (raw)
DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation
Yexi Chen et al. Genes Dev. 2009.
Abstract
Transcription elongation factor DSIF/Spt4-Spt5 is capable of promoting and inhibiting RNA polymerase II elongation and is involved in the expression of various genes. While it has been known for many years that DSIF inhibits elongation in collaboration with the negative elongation factor NELF, how DSIF promotes elongation is largely unknown. Here, an activity-based biochemical approach was taken to understand the mechanism of elongation activation by DSIF. We show that the Paf1 complex (Paf1C) and Tat-SF1, two factors implicated previously in elongation control, collaborate with DSIF to facilitate efficient elongation. In human cells, these factors are recruited to the FOS gene in a temporally coordinated manner and contribute to its high-level expression. We also show that elongation activation by these factors depends on P-TEFb-mediated phosphorylation of the Spt5 C-terminal region. A clear conclusion emerging from this study is that a set of elongation factors plays nonredundant, cooperative roles in elongation. This study also shows unambiguously that Paf1C, which is generally thought to have chromatin-related functions, is involve directlyd in elongation control.
Figures
Figure 1.
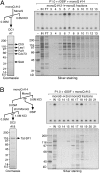
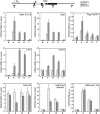
Identification of DC activity. (A) rDSIF promotes transcription elongation in HeLa NE. Transcription reactions contained the template pSLG402 (shown schematically to the right) and either NE or NE devoid of DSIF (NEΔ). Transcription was allowed to proceed for the indicated times. (MLP) Adenovirus major-late promoter. (B) rDSIF does not promote transcription elongation in P1.0. Phosphocellulose fractions (see Fig. 2A) were used instead of NE. “Δ” indicates the fractions that were immunodepleted with anti-Spt5 antibody and were hence devoid of DSIF. (C) Immunoblot analysis of the phosphocellulose fractions. (D) P.3-derived DEAE Sepharose fractions (see Fig. 2A) were assayed for DC activity. (E) Immunoblot analysis of the DEAE Sepharose fractions.
Figure 2.
Separation of DC activity into two components. (A) Scheme for partial purification of DCs. (B–D) D.3-derived mono-Q fractions were assayed for DC activity. In B, P1.0, rDSIF, and one of the column fractions were used as protein source. An anti-Spt5 immunoblot is shown below. In C and D, the same mono-Q fractions were assayed in the presence of an additional mono-Q fraction.
Figure 3.
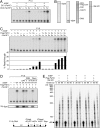
Final purification and identification of DCs. (A) Final purification of DC1 was performed according to the scheme shown. Mono-S fractions were assayed for DC1 activity in the presence of P1.0, rDSIF, and crude DC2 (mono-Q #14). The same fractions were analyzed by SDS-PAGE and silver staining. Proteins in an active fraction were also visualized by Coomassie staining and were subjected to LC-MS/MS analysis. (B) Final purification of DC2 was performed according to the scheme shown. Fractions derived from the second mono-Q column were assayed for DC2 activity in the presence of P1.0, rDSIF, and crude DC1 (mono-Q #12). Proteins were analyzed as described in A.
Figure 4.
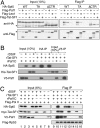
Cooperative action of DSIF, Paf1C, and Tat-SF1 on elongation. (A) Transcription assays were carried out in the presence of various combinations of rDSIF and DCs, each from the final purification step. (B) Purified proteins used for in vitro transcription. (C) Transcription assays were carried out in the presence of various combinations of rDSIF, Flag-Paf1C, and rTat-SF1. Below, percent read-through was calculated from relative intensity of the two products by taking into account the number of uridines (labeled nucleotides) in each fragment. (D) Requirement for P-TEFb-mediated phosphorylation of the Spt5 CTR in productive elongation. rDSIF composed of rSpt4 and wild-type rSpt5 or one of its mutants was used in combination with Flag-Paf1C and rTat-SF1. Where indicated, P-TEFb inhibitors such as DRB and Flavopiridol were included. In the top panel, transcription reactions were performed in the presence of [α-32P] UTP, whereas in the bottom panel, an identical set of reactions were performed in the presence of [γ-32P] ATP, followed by immunoprecipitation of Spt5. (E) Pulse-chase transcription assays were carried out as described in the Materials and Methods. The indicated combinations of elongation factors (ECs) were added to reactions 10 min prior to chase.
Figure 5.
Physical interactions of DSIF, Paf1C, Tat-SF1, and Pol II. (A) Coimmunoprecipitation of transiently expressed proteins. Following transfection of the indicated constructs into HeLa cells, Flag-tagged proteins were immunopurified from cell extracts using anti-Flag M2 agarose and then immunoblotted with the indicated antibodies. Asterisks denote nonspecific signal. (B) Coimmunoprecipitation was carried out using 100 ng each of rDSIF containing HA-Spt5, rTat-SF1, and rPaf1C containing V5-Paf1 that were individually expressed and purified from Sf-9 cells. (C) NE prepared from HeLa/FH3 cells (Hasegawa et al. 2003) stably expressing the Flag- and His-tagged Rpb3 subunit of Pol II was incubated with anti-Flag M2 agarose to immobilize 1 μg of FH-Pol II to the beads. The Pol II beads were then incubated with rPaf1C (2 μg), rTat-SF1 (2 μg), and rDSIF (2 μg) in various combinations for 2 h at 4°C and washed several times with NP-40 buffer (50 mM Tris at pH 7.9, 150 mM NaCl, 1% NP-40). Bound materials were eluted with Flag peptide and subjected to immunoblotting.
Figure 6.
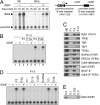
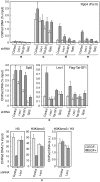
Nonredundant roles of DSIF, Paf1C, and Tat-SF1 in EGF-induced transcription. (A) HeLa cells were transduced with recombinant lentivirus expressing no shRNA (Control) or expressing one of the shRNAs. Four days post-infection, immunoblotting was performed. (B) Four days to 6 d post-infection, the cells were treated with EGF for the indicated times and then harvested for RNA analysis. The expression levels of the FOS and EGR1 mRNAs were quantified by real-time RT–PCR, and the values are expressed as fold changes from the mRNA levels in control virus-transduced, unstimulated cells.
Figure 7.
Concomitant association of DSIF, Paf1C, Tat-SF1, and Pol II with the FOS gene. ChIP assays were carried out as described in the Materials and Methods. Prior to harvest, HeLa cells were cultured for 18 h in the presence of 0.2% serum and then stimulated for 7.5 min with 0.1 mg/mL EGF or were left untreated. The amplicons used are presented as bars below the diagram of the FOS gene. All of the data are means ± SEM from three independent experiments and are expressed as percent input. Enrichment ratios of histone H3K4me3 to total histone H3 were calculated from respective ChIP data and are expressed in arbitrary units.
Figure 8.
Cooperative assembly of DSIF, Paf1, and Tat-SF1 into the Pol II elongation complex. HeLa cells were transduced with recombinant lentivirus expressing no shRNA (Control), Leo1 shRNA #1, Tat-SF1 shRNA #1, or Spt5 shRNA. Five days to 6 d post-infection, the cells were treated with EGF for 7.5 min or were left untreated and subjected to ChIP. Data are means ± SEM from three independent experiments. Anti-H3K4me3 from Upstate Biotechnologies (07-473) was used where indicated.
Similar articles
- Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II.
Lu X, Zhu X, Li Y, Liu M, Yu B, Wang Y, Rao M, Yang H, Zhou K, Wang Y, Chen Y, Chen M, Zhuang S, Chen LF, Liu R, Chen R. Lu X, et al. Nucleic Acids Res. 2016 Aug 19;44(14):6853-67. doi: 10.1093/nar/gkw571. Epub 2016 Jun 28. Nucleic Acids Res. 2016. PMID: 27353326 Free PMC article. - DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs.
Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. Wada T, et al. Genes Dev. 1998 Feb 1;12(3):343-56. doi: 10.1101/gad.12.3.343. Genes Dev. 1998. PMID: 9450929 Free PMC article. - Mechanisms of Transcription Elongation Factor DSIF (Spt4-Spt5).
Decker TM. Decker TM. J Mol Biol. 2021 Jul 9;433(14):166657. doi: 10.1016/j.jmb.2020.09.016. Epub 2020 Sep 25. J Mol Biol. 2021. PMID: 32987031 Review. - The pleiotropic roles of SPT5 in transcription.
Song A, Chen FX. Song A, et al. Transcription. 2022 Feb-Jun;13(1-3):53-69. doi: 10.1080/21541264.2022.2103366. Epub 2022 Jul 25. Transcription. 2022. PMID: 35876486 Free PMC article. Review.
Cited by
- The Paf1 complex and P-TEFb have reciprocal and antagonist roles in maintaining multipotent neural crest progenitors.
Jurynec MJ, Bai X, Bisgrove BW, Jackson H, Nechiporuk A, Palu RAS, Grunwald HA, Su YC, Hoshijima K, Yost HJ, Zon LI, Grunwald DJ. Jurynec MJ, et al. Development. 2019 Dec 16;146(24):dev180133. doi: 10.1242/dev.180133. Development. 2019. PMID: 31784460 Free PMC article. - Cis and trans interactions between genes encoding PAF1 complex and ESCRT machinery components in yeast.
Rodrigues J, Lydall D. Rodrigues J, et al. Curr Genet. 2018 Oct;64(5):1105-1116. doi: 10.1007/s00294-018-0828-6. Epub 2018 Mar 22. Curr Genet. 2018. PMID: 29564528 Free PMC article. - Cell-cell adhesion regulates Merlin/NF2 interaction with the PAF complex.
Roehrig AE, Klupsch K, Oses-Prieto JA, Chaib S, Henderson S, Emmett W, Young LC, Surinova S, Blees A, Pfeiffer A, Tijani M, Brunk F, Hartig N, Muñoz-Alegre M, Hergovich A, Jennings BH, Burlingame AL, Rodriguez-Viciana P. Roehrig AE, et al. PLoS One. 2021 Aug 23;16(8):e0254697. doi: 10.1371/journal.pone.0254697. eCollection 2021. PLoS One. 2021. PMID: 34424918 Free PMC article. - Pathogenesis of Börjeson-Forssman-Lehmann syndrome: Insights from PHF6 function.
Jahani-Asl A, Cheng C, Zhang C, Bonni A. Jahani-Asl A, et al. Neurobiol Dis. 2016 Dec;96:227-235. doi: 10.1016/j.nbd.2016.09.011. Epub 2016 Sep 12. Neurobiol Dis. 2016. PMID: 27633282 Free PMC article. Review. - The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin.
Yu X, Michaels SD. Yu X, et al. Plant Physiol. 2010 Jul;153(3):1074-84. doi: 10.1104/pp.110.158386. Epub 2010 May 12. Plant Physiol. 2010. PMID: 20463090 Free PMC article.
References
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases