Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks - PubMed (original) (raw)
. 2009 Aug 26;4(8):e6789.
doi: 10.1371/journal.pone.0006789.
Alex J Kuo, Peggie Cheung, Chih Long Liu, Valentina Migliori, Alexsandra Espejo, Fabio Casadio, Christian Bassi, Bruno Amati, Mark T Bedford, Ernesto Guccione, Or Gozani
Affiliations
- PMID: 19956676
- PMCID: PMC2777412
- DOI: 10.1371/journal.pone.0006789
Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks
Dennis J Bua et al. PLoS One. 2009.
Abstract
Knowledge of protein domains that function as the biological effectors for diverse post-translational modifications of histones is critical for understanding how nuclear and epigenetic programs are established. Indeed, mutations of chromatin effector domains found within several proteins are associated with multiple human pathologies, including cancer and immunodeficiency syndromes. To date, relatively few effector domains have been identified in comparison to the number of modifications present on histone and non-histone proteins. Here we describe the generation and application of human modified peptide microarrays as a platform for high-throughput discovery of chromatin effectors and for epitope-specificity analysis of antibodies commonly utilized in chromatin research. Screening with a library containing a majority of the Royal Family domains present in the human proteome led to the discovery of TDRD7, JMJ2C, and MPP8 as three new modified histone-binding proteins. Thus, we propose that peptide microarray methodologies are a powerful new tool for elucidating molecular interactions at chromatin.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
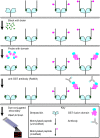
Figure 1. Key steps in the human epigenome peptide microarray (HEMP) procedure.
Figure 2. HEMP slides as a diagnostic tool for testing antibody specificity.
(a) Array images for antibodies: i) anti-H3K9me1, ii) anti-H3R2me2 (asymmetric), iii) anti-H3K18ac, iv) anti-γH2AX, and v) anti-H3K20me3 with schematic of array layout and key. (b) Heatmap representation of antibody HEMP slide data (See Table S2 for additional antibody details). The epitope(s) that the antibody was generated against is/are highlighted with a white border. See “Heatmap PTM key” for details about peptides with post-translational modifications (PTMs). Note, that di-methyl arginine residues with blue circles are symmetrically di-methylated. SNR RN, signal-to-noise ratio range-normalized. n.t., not tested.
Figure 3. Detection of known chromatin effector-histone PTM interactions using HEMP slides.
(a) The chromodomain of Drosophila melanogaster heterochromatin protein 1 alpha (dsHP1CD), (b) the plant homeodomain of human inhibitor of growth 3 (ING3PHD), and (c) the double chromodomains of human chromodomain-helicase-DNA-binding protein 1 (CHD1CD), all recognize, as indicated, their cognate histone ligand on the peptide array. All these protein domains are expressed as GST-fusions and an array probed with GST alone (d) serves as a negative control. For order of peptide spotting, see schematic in Figure 2a.
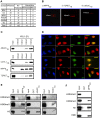
Figure 4. Identification of three novel methyl-histone binding modules.
(a) Table summarizing the number of domains tested in this study and the number of interactions detected. CD, chromodomain. TD, tudor domain. (b) Array images for: i) MPP8CD, ii) TDRD7TD, and iii) JMJ2CTD. Peptide/s detected in each experiment is indicated. See Figure 2a for array schematic. (c) Validation of array results in peptide-binding assays. Biotinylated peptide pull-down assay using peptides detected in (b) and the indicated GST-fusion proteins. (d) Co-localization of CDYL1 with H3K9me3. Representative immunofluorescence images of U2OS cells transfected with His-tagged CDYL1 and co-stained with the indicated antibodies. K9 = H3K9me3, K4 = H3K4me3. (e) Validation of array results in bulk-histone binding assays. Calf-thymus histone pull-down with the indicated proteins: MPP8CD, TDRD7TD, and JMJ2CTD. In each case the domain was pulled-down and the pellet was probed with the indicated antibodies. (f) MPP8CD binds to HeLa-purified nucleosomes enriched for H3K9me3 but not H3K4me3. Pull-downs of GST or GST-MPP8CD protein after incubation with HeLa nucleosomes were probed with the antibodies indicated (see Fig. S1 for quantitation).
Similar articles
- Application of Celluspots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails.
Bock I, Kudithipudi S, Tamas R, Kungulovski G, Dhayalan A, Jeltsch A. Bock I, et al. BMC Biochem. 2011 Aug 31;12:48. doi: 10.1186/1471-2091-12-48. BMC Biochem. 2011. PMID: 21884582 Free PMC article. - A SPOT on the chromatin landscape? Histone peptide arrays as a tool for epigenetic research.
Nady N, Min J, Kareta MS, Chédin F, Arrowsmith CH. Nady N, et al. Trends Biochem Sci. 2008 Jul;33(7):305-13. doi: 10.1016/j.tibs.2008.04.014. Epub 2008 Jun 4. Trends Biochem Sci. 2008. PMID: 18538573 - High-throughput profiling of histone post-translational modifications and chromatin modifying proteins by reverse phase protein array.
Wang X, Shi Z, Lu HY, Kim JJ, Bu W, Villalobos JA, Perera DN, Jung SY, Wang T, Grimm SL, Taylor BC, Rajapakshe K, Park H, Wulfkuhle J, Young NL, Li Y, Coarfa C, Edwards DP, Huang S. Wang X, et al. J Proteomics. 2022 Jun 30;262:104596. doi: 10.1016/j.jprot.2022.104596. Epub 2022 Apr 27. J Proteomics. 2022. PMID: 35489683 Free PMC article. - Engineering chromatin states: chemical and synthetic biology approaches to investigate histone modification function.
Pick H, Kilic S, Fierz B. Pick H, et al. Biochim Biophys Acta. 2014 Aug;1839(8):644-56. doi: 10.1016/j.bbagrm.2014.04.016. Epub 2014 Apr 21. Biochim Biophys Acta. 2014. PMID: 24768924 Review. - Application of modified histone peptide arrays in chromatin research.
Mauser R, Jeltsch A. Mauser R, et al. Arch Biochem Biophys. 2019 Jan;661:31-38. doi: 10.1016/j.abb.2018.10.019. Epub 2018 Nov 2. Arch Biochem Biophys. 2019. PMID: 30391375 Review.
Cited by
- Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity.
Ballaré C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, Carlomagno T, Benitah SA, Di Croce L. Ballaré C, et al. Nat Struct Mol Biol. 2012 Dec;19(12):1257-65. doi: 10.1038/nsmb.2434. Epub 2012 Oct 28. Nat Struct Mol Biol. 2012. PMID: 23104054 Free PMC article. - RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36.
Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. Fuchs SM, et al. J Biol Chem. 2012 Jan 27;287(5):3249-56. doi: 10.1074/jbc.M111.273953. Epub 2011 Dec 7. J Biol Chem. 2012. PMID: 22157004 Free PMC article. - Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns.
Nikolov M, Stützer A, Mosch K, Krasauskas A, Soeroes S, Stark H, Urlaub H, Fischle W. Nikolov M, et al. Mol Cell Proteomics. 2011 Nov;10(11):M110.005371. doi: 10.1074/mcp.M110.005371. Epub 2011 Aug 11. Mol Cell Proteomics. 2011. PMID: 21836164 Free PMC article. - A PWWP Domain of Histone-Lysine N-Methyltransferase NSD2 Binds to Dimethylated Lys-36 of Histone H3 and Regulates NSD2 Function at Chromatin.
Sankaran SM, Wilkinson AW, Elias JE, Gozani O. Sankaran SM, et al. J Biol Chem. 2016 Apr 15;291(16):8465-74. doi: 10.1074/jbc.M116.720748. Epub 2016 Feb 24. J Biol Chem. 2016. PMID: 26912663 Free PMC article. - Keep quiet: the HUSH complex in transcriptional silencing and disease.
Müller I, Helin K. Müller I, et al. Nat Struct Mol Biol. 2024 Jan;31(1):11-22. doi: 10.1038/s41594-023-01173-7. Epub 2024 Jan 12. Nat Struct Mol Biol. 2024. PMID: 38216658 Review.
References
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. - PubMed
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22(9):836–845. - PubMed
- Bannister AJ, Kouzarides T. Histone methylation: Recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. - PubMed
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111(6):771–778. - PubMed
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources