High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains - PubMed (original) (raw)
. 2010 Mar 1;9(2):129-59.
doi: 10.1111/j.1601-183X.2009.00540.x. Epub 2009 Sep 22.
S Duvvuru, B Gomero, T A Ansah, C D Blaha, M N Cook, K M Hamre, W R Lariviere, D B Matthews, G Mittleman, D Goldowitz, E J Chesler
Affiliations
- PMID: 19958391
- PMCID: PMC2855868
- DOI: 10.1111/j.1601-183X.2009.00540.x
Free PMC article
High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains
V M Philip et al. Genes Brain Behav. 2010.
Free PMC article
Abstract
Genetic reference populations, particularly the BXD recombinant inbred (BXD RI) strains derived from C57BL/6J and DBA/2J mice, are a valuable resource for the discovery of the bio-molecular substrates and genetic drivers responsible for trait variation and covariation. This approach can be profitably applied in the analysis of susceptibility and mechanisms of drug and alcohol use disorders for which many predisposing behaviors may predict the occurrence and manifestation of increased preference for these substances. Many of these traits are modeled by common mouse behavioral assays, facilitating the detection of patterns and sources of genetic coregulation of predisposing phenotypes and substance consumption. Members of the Tennessee Mouse Genome Consortium (TMGC) have obtained phenotype data from over 250 measures related to multiple behavioral assays across several batteries: response to, and withdrawal from cocaine, 3,4-methylenedioxymethamphetamine; "ecstasy" (MDMA), morphine and alcohol; novelty seeking; behavioral despair and related neurological phenomena; pain sensitivity; stress sensitivity; anxiety; hyperactivity and sleep/wake cycles. All traits have been measured in both sexes in approximately 70 strains of the recently expanded panel of BXD RI strains. Sex differences and heritability estimates were obtained for each trait, and a comparison of early (N = 32) and recent (N = 37) BXD RI lines was performed. Primary data are publicly available for heritability, sex difference and genetic analyses using the MouseTrack database, and are also available in GeneNetwork.org for quantitative trait locus (QTL) detection and genetic analysis of gene expression. Together with the results of related studies, these data form a public resource for integrative systems genetic analysis of neurobehavioral traits.
Figures
Figure 1
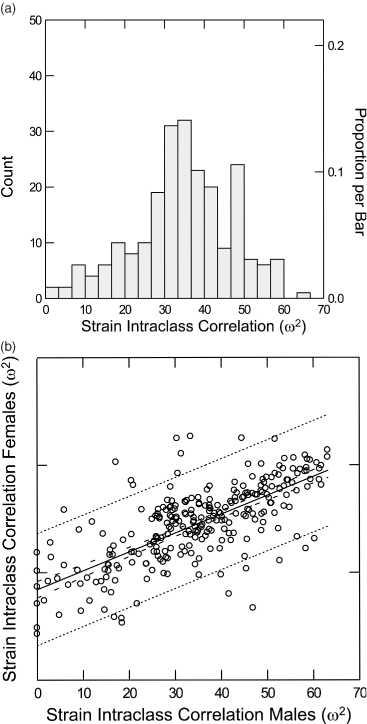
Strain intra-class correlations for all measures. (a) Frequency histogram of strain intra-class correlations for both sexes combined. A majority of the behavioral measures (146 of 257) have ω2 greater than 30% making them amenable to QTL mapping. (b) Scatter plot of strain intra-class correlations for males and females. Dotted lines are 95% upper and lower prediction intervals for the relationship between these values. Dashed lines are the corresponding 95% upper and lower confidence intervals.
Figure 3
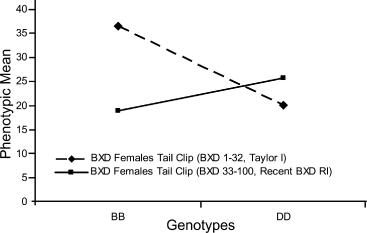
Effect plot of marker gnf01.018.340 (Chr 1 at 21.43 Mb) in Taylor I (BXD 1–32) and Recent RI (BXD 33–100). The marker present at the suggestive QTL in Taylor 1 has a higher phenotypic means for the B allele when compared with the D allele. This difference is significant and therefore results in the presence of a QTL. The same marker in the Recent BXD RI lines has a higher phenotypic mean for the D allele, but no QTL is present within this strain set.
Figure 2
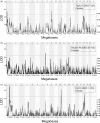
Quantitative trait locus analysis of mechanical nociception in the original and recent BXD RI panels: QTL analysis of a single-trait, the latency to respond to a plastic-coated smooth alligator clip placed on the tail was performed across three BXD panels, namely, Taylor I (BXD 1–32), Recent BXD RI (BXD 33–100) and Full BXD RI (BXD 1–100). In females, suggestive QTL were detected on Chr 1 and 9 for Taylor I (a), Chr 8 for the Recent BXD RI (BXD 33–100) (b) and Chr 1, 11 and 17 for the Full RI Lines (c). Mapping of tail clip latency in males showed no suggestive QTL for Taylor I (d), suggestive QTL on Chr 2, 9 and 18 for Recent BXD RI (BXD 33–100) (e), and two suggestive QTL on Chr 2 and 9 for the Full RI panel (f).
Figure 2
Quantitative trait locus analysis of mechanical nociception in the original and recent BXD RI panels: QTL analysis of a single-trait, the latency to respond to a plastic-coated smooth alligator clip placed on the tail was performed across three BXD panels, namely, Taylor I (BXD 1–32), Recent BXD RI (BXD 33–100) and Full BXD RI (BXD 1–100). In females, suggestive QTL were detected on Chr 1 and 9 for Taylor I (a), Chr 8 for the Recent BXD RI (BXD 33–100) (b) and Chr 1, 11 and 17 for the Full RI Lines (c). Mapping of tail clip latency in males showed no suggestive QTL for Taylor I (d), suggestive QTL on Chr 2, 9 and 18 for Recent BXD RI (BXD 33–100) (e), and two suggestive QTL on Chr 2 and 9 for the Full RI panel (f).
Figure 4
Factor scores from individual strains plotted on a set of axes each representing a factor show a behavioral profile of each strain. Although some strains exhibit different magnitudes of similar profiles, others are distinct. Axes labels represent the first eight factors: (1) injection stress-induced locomotor activity, (2) morphine withdrawal (3) morphine response, (4) reactivity, (5) locomotor activity in a novel environment, (6) conflict avoidance, (7) morphine activity duration and (8) cocaine sensitization. All strain profiles are available in supplementary figure.
Figure 5
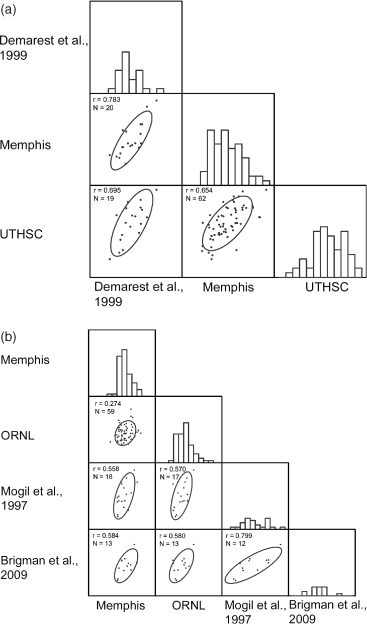
Across trait and across studies correlations: (a) saline-induced locomotor activity: in our study two laboratories, Memphis (Cocaine) and UTHSC (Ethanol), collected data on open field locomotion following saline in 15 and 20 min on 64 BXD RI strains, respectively. We compared these data with data collected on saline-induced locomotor response by Demarest et al. (1999) on 25 BXD RI strains. Results indicate that there is good correlation among data on saline-induced locomotor response across the laboratories and to a previously published study. Correlations among the r_Memphis,UTHSC = 0.556,r_Memphis,Demarest = 0.781 and _r_Demarest,UTHSC = 0.695. (b) Thermal nociception: thermal nociception (hot plate latency) was preformed in two laboratories, namely, ORNL and Memphis in this study. Data collected in this study were compared with a previously published study of thermal nociception by Mogil et al. (1997) and Brigman et al. (2009). Correlations ranged from 0.274 to 0.799 (Fig. 4b). The low correlation among the data collected at the Memphis and ORNL laboratories may be attributed to the two different hot plate temperatures used.
Similar articles
- Systems genetics of sensation seeking.
Dickson PE, Roy TA, McNaughton KA, Wilcox TD, Kumar P, Chesler EJ. Dickson PE, et al. Genes Brain Behav. 2019 Mar;18(3):e12519. doi: 10.1111/gbb.12519. Epub 2018 Oct 8. Genes Brain Behav. 2019. PMID: 30221471 Free PMC article. - Using gene expression databases for classical trait QTL candidate gene discovery in the BXD recombinant inbred genetic reference population: mouse forebrain weight.
Lu L, Wei L, Peirce JL, Wang X, Zhou J, Homayouni R, Williams RW, Airey DC. Lu L, et al. BMC Genomics. 2008 Sep 25;9:444. doi: 10.1186/1471-2164-9-444. BMC Genomics. 2008. PMID: 18817551 Free PMC article. - Identifying genetic loci and spleen gene coexpression networks underlying immunophenotypes in BXD recombinant inbred mice.
Lynch RM, Naswa S, Rogers GL Jr, Kania SA, Das S, Chesler EJ, Saxton AM, Langston MA, Voy BH. Lynch RM, et al. Physiol Genomics. 2010 May;41(3):244-53. doi: 10.1152/physiolgenomics.00020.2010. Epub 2010 Feb 23. Physiol Genomics. 2010. PMID: 20179155 Free PMC article. - Use of recombinant inbred strains to identify quantitative trait loci in psychopharmacology.
Gora-Maslak G, McClearn GE, Crabbe JC, Phillips TJ, Belknap JK, Plomin R. Gora-Maslak G, et al. Psychopharmacology (Berl). 1991;104(4):413-24. doi: 10.1007/BF02245643. Psychopharmacology (Berl). 1991. PMID: 1780413 Review. - BXD Recombinant Inbred Mice as a Model to Study Neurotoxicity.
Martins AC, López-Granero C, Ferrer B, Tinkov AA, Skalny AV, Paoliello MMB, Aschner M. Martins AC, et al. Biomolecules. 2021 Nov 25;11(12):1762. doi: 10.3390/biom11121762. Biomolecules. 2021. PMID: 34944406 Free PMC article. Review.
Cited by
- Neuroinflammation, Pain and Depression: An Overview of the Main Findings.
Campos ACP, Antunes GF, Matsumoto M, Pagano RL, Martinez RCR. Campos ACP, et al. Front Psychol. 2020 Jul 31;11:1825. doi: 10.3389/fpsyg.2020.01825. eCollection 2020. Front Psychol. 2020. PMID: 32849076 Free PMC article. Review. - Systems genetics of sensation seeking.
Dickson PE, Roy TA, McNaughton KA, Wilcox TD, Kumar P, Chesler EJ. Dickson PE, et al. Genes Brain Behav. 2019 Mar;18(3):e12519. doi: 10.1111/gbb.12519. Epub 2018 Oct 8. Genes Brain Behav. 2019. PMID: 30221471 Free PMC article. - Heritable variation in locomotion, reward sensitivity and impulsive behaviors in a genetically diverse inbred mouse panel.
Bailey LS, Bagley JR, Dodd R, Olson A, Bolduc M, Philip VM, Reinholdt LG, Sukoff Rizzo SJ, Tarantino L, Gagnon L, Chesler EJ, Jentsch JD. Bailey LS, et al. Genes Brain Behav. 2021 Nov;20(8):e12773. doi: 10.1111/gbb.12773. Epub 2021 Oct 21. Genes Brain Behav. 2021. PMID: 34672075 Free PMC article. - Initial genetic dissection of serum neuroactive steroids following chronic intermittent ethanol across BXD mouse strains.
Porcu P, O'Buckley TK, Lopez MF, Becker HC, Miles MF, Williams RW, Morrow AL. Porcu P, et al. Alcohol. 2017 Feb;58:107-125. doi: 10.1016/j.alcohol.2016.07.011. Epub 2016 Nov 10. Alcohol. 2017. PMID: 27884493 Free PMC article. - Post-genomic behavioral genetics: From revolution to routine.
Ashbrook DG, Mulligan MK, Williams RW. Ashbrook DG, et al. Genes Brain Behav. 2018 Mar;17(3):e12441. doi: 10.1111/gbb.12441. Epub 2017 Dec 21. Genes Brain Behav. 2018. PMID: 29193773 Free PMC article. Review.
References
- Bear B, Asgian J, Termin A, Zimmermann N. Small molecules targeting sodium and calcium channels for neuropathic pain. Curr Opin Drug Discov Devel. 2009;12:543–561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AA016662/AA/NIAAA NIH HHS/United States
- U01 AA013509/AA/NIAAA NIH HHS/United States
- DA020677/DA/NIDA NIH HHS/United States
- R01 DA020677/DA/NIDA NIH HHS/United States
- AA016662/AA/NIAAA NIH HHS/United States
- R01 DA021198/DA/NIDA NIH HHS/United States
- U01 AA016669-03/AA/NIAAA NIH HHS/United States
- U01 AA013499/AA/NIAAA NIH HHS/United States
- U01 AA013509-06/AA/NIAAA NIH HHS/United States
- AA13509/AA/NIAAA NIH HHS/United States
- U01 AA016669/AA/NIAAA NIH HHS/United States
- R01 DA020677-04/DA/NIDA NIH HHS/United States
- DA021198/DA/NIDA NIH HHS/United States
- AA13499/AA/NIAAA NIH HHS/United States
- U01 AA016662-03/AA/NIAAA NIH HHS/United States
- R01 DA021198-03/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases