PcG recruitment by the YY1 REPO domain can be mediated by Yaf2 - PubMed (original) (raw)
PcG recruitment by the YY1 REPO domain can be mediated by Yaf2
Frank Wilkinson et al. J Cell Biochem. 2010.
Abstract
The Polycomb Group (PcG) complex of transcriptional repressors is critical for the maintenance of stage-specific developmental gene expression, stem cell maintenance and for large-scale chromosomal dynamics. Functional deficiency of a single PcG gene can severely compromise PcG function, leading to developmental defects, embryonic lethality, or a number of malignancies. Despite the critical nature of PcG proteins, the mechanisms by which these complexes mediate their effects are relatively uncharacterized. Nearly all vertebrate PcG proteins lack inherent DNA binding capacity, making it unclear how they are targeted to Polycomb response element (PRE) sequences. Transcription factor YY1 is a functional ortholog of a Drosophila PcG protein, Pleiohomeotic (PHO), one of the few PcG proteins with specific DNA binding capability, and YY1 can recruit PcG proteins to specific DNA sequences. A small 25 amino acid YY1 domain (the REPO domain) is necessary and sufficient for recruitment of PcG proteins to DNA and for transcriptional repression. We show here that the YY1 REPO domain interacts with PcG protein Yaf2 and recruits Yaf2 to DNA. Interaction is lost when the YY1 REPO domain is deleted. In addition we show that Yaf2, when linked to a heterologous DNA binding domain, can recruit PcG proteins to DNA leading to transcriptional repression. When the Drosophila homolog of Yaf2 (dRYBP) is mutated, PcG recruitment to DNA is reduced. Taken together, our results suggest that Yaf2 serves as a molecular bridge between YY1 and other PcG complex proteins.
(c) 2009 Wiley-Liss, Inc.
Figures
Fig. 1
Yeast two hybrid detection of REPO–Yaf2 interactions. S. cerevisiae AH109 was transformed with the indicated bait and prey constructs. Cotransformants were passaged onto selective medium (Trp/Leu/Ade/His dropout medium). The plate on the left contains pGADt7 empty vector expressing the GAL4 activation domain (AD) and the plate on the right contains pGADt7 vector expressing the AD fused to full-length Yaf2 (residues 1–179, pGADt7-Yaf2). Bait constructs expressing GAL4 DBD fusions are as indicated: BK, pGBKt7 empty vector control; YY1, full-length YY1 1–414; YY1 1–414Δ201–226; YY1 201–226 REPO, YY1 residues 201–226. The scheme below diagrams the strategy used in the assay. Transcription of the nutritional markers HIS3 and ADE2 from interactions resulting from bait-prey binding interactions (two head arrow) is indicated.
Fig. 2
Yaf2 is bound to DNA in the presence of GALREPO. A: Diagram of the BGUZ transgenic reporter. The reporter consists of the LacZ coding sequence under control of the Ubx promoter and BXD enhancer elements. The multi-merized Gal4 recognition sequence is upstream from the Ubx promoter. The black arrows indicate the approximate positions of the PCR primers used in panel B. B: Agarose gel electrophoresis of PCR products detected from ChIP assays stained with ethidium bromide. The cross indicating the chromatin source is indicated on the left. The triangles indicate a 10-fold change in template concentration. Antibodies used for the immunoprecipitations are indicated above the appropriate lanes. M indicates molecular weight markers. Numbers indicate lanes referred to in the text.
Fig. 3
Yaf2 recruits PcG proteins to DNA. Agarose gel electrophoresis of PCR products detected by ChIP assay stained with ethidium bromide. The BGUZ reporter and PCR primer locations are as indicated in Figure 2. The cross indicating the chromatin source is indicated on the left. The triangles indicate a 10-fold change in template concentration. Antibodies used for the immunoprecipitation are indicated above the appropriate lanes. M indicates molecular weight markers. Numbers indicate lanes referred to in the text.
Fig. 4
Transcriptional silencing of BGUZ by GALYaf2. A: BGUZ reporter transgene, hunchback-GALYaf2 effector transgene, and expression patterns of BGUZ, hunchback-GALYaf2, and the superimposed expression patterns in embryos. B: Drosophila embryos resulting from crosses with BGUZ females (1 h timed egg lay) fixed and stained with X-gal at hour 6. Crosses are indicated above each panel: BGUZ, BGUZ females × BGUZ males; other panels, BGUZ females crossed to distinct transgenic hunchback-GALYaf2 males.
Fig. 5
dRYBP mutation reduced PcG binding to PRED. Agarose gel electrophoresis of PCR products detected by ChIP assay stained with ethidium bromide. The strain indicating the chromatin source is indicated on the left. The triangles indicate a 10-fold change in template concentration. Antibodies used for the immunoprecipitation are indicated above the appropriate lanes. M indicates molecular weight markers. Numbers indicate lanes referred to in the text.
Fig. 6
Yeast two hybrid mapping of interactions between Yaf2 fragments and the YY1 REPO domain. S. cerevisiae AH109 was transformed with the indicated bait and prey constructs. Cotransformants were passaged onto selective medium (Trp/Leu/Ade/His dropout medium). The streaked cultures on the left contain pGADt7 expressing the GAL4 activation domain (AD) fused to Yaf2 residues 1–101 (AD-N) and the streaked cultures on the right contain pGADt7 expressing the AD fused to Yaf2 residues 102–179 (AD-C). These were cotransformed with bait constructs pGBKt7 empty vector (BK) or pGBKt7 expressing the GAL4 DBD fused to YY1 201–226 (YY1 REPO). Transcription of the nutritional markers HIS3 and ADE2 from interactions resulting from bait–prey binding interactions is as indicated in Figure 1. [Color figure can be viewed in the online issue, which is available at
.]
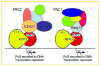
Fig. 7
Model of YY1 REPO–Yaf2–PcG interactions. YY1 bound to DNA is shown with the REPO domain interacting with Yaf2, and Yaf2 interacting with either PRC2 or PRC1. Candidate PRC2 and PRC1 interacting proteins EZH2 and Ring-1 are indicated respectively. [Color figure can be viewed in the online issue, which is available at
.]
Similar articles
- YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment.
Basu A, Wilkinson FH, Colavita K, Fennelly C, Atchison ML. Basu A, et al. Nucleic Acids Res. 2014 Feb;42(4):2208-23. doi: 10.1093/nar/gkt1187. Epub 2013 Nov 26. Nucleic Acids Res. 2014. PMID: 24285299 Free PMC article. - Polycomb recruitment to DNA in vivo by the YY1 REPO domain.
Wilkinson FH, Park K, Atchison ML. Wilkinson FH, et al. Proc Natl Acad Sci U S A. 2006 Dec 19;103(51):19296-301. doi: 10.1073/pnas.0603564103. Epub 2006 Dec 8. Proc Natl Acad Sci U S A. 2006. PMID: 17158804 Free PMC article. - Structural basis for targeting the chromatin repressor Sfmbt to Polycomb response elements.
Alfieri C, Gambetta MC, Matos R, Glatt S, Sehr P, Fraterman S, Wilm M, Müller J, Müller CW. Alfieri C, et al. Genes Dev. 2013 Nov 1;27(21):2367-79. doi: 10.1101/gad.226621.113. Genes Dev. 2013. PMID: 24186981 Free PMC article. - Polycomb response elements and targeting of Polycomb group proteins in Drosophila.
Müller J, Kassis JA. Müller J, et al. Curr Opin Genet Dev. 2006 Oct;16(5):476-84. doi: 10.1016/j.gde.2006.08.005. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16914306 Review. - Chromatin regulation: how complex does it get?
Meier K, Brehm A. Meier K, et al. Epigenetics. 2014 Nov;9(11):1485-95. doi: 10.4161/15592294.2014.971580. Epigenetics. 2014. PMID: 25482055 Free PMC article. Review.
Cited by
- Assessing the dynamics and macromolecular interactions of the intrinsically disordered protein YY1.
Donald H, Blane A, Buthelezi S, Naicker P, Stoychev S, Majakwara J, Fanucchi S. Donald H, et al. Biosci Rep. 2023 Oct 31;43(10):BSR20231295. doi: 10.1042/BSR20231295. Biosci Rep. 2023. PMID: 37815922 Free PMC article. - The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability.
Fitieh A, Locke AJ, Motamedi M, Ismail IH. Fitieh A, et al. Int J Mol Sci. 2021 Mar 15;22(6):2976. doi: 10.3390/ijms22062976. Int J Mol Sci. 2021. PMID: 33804165 Free PMC article. Review. - Roles Played by YY1 in Embryonic, Adult and Cancer Stem Cells.
Martinez-Ruiz GU, Morales-Sanchez A, Pacheco-Hernandez AF. Martinez-Ruiz GU, et al. Stem Cell Rev Rep. 2021 Oct;17(5):1590-1606. doi: 10.1007/s12015-021-10151-9. Epub 2021 Mar 17. Stem Cell Rev Rep. 2021. PMID: 33728560 Free PMC article. Review. - The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1.
Verheul TCJ, van Hijfte L, Perenthaler E, Barakat TS. Verheul TCJ, et al. Front Cell Dev Biol. 2020 Sep 30;8:592164. doi: 10.3389/fcell.2020.592164. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33102493 Free PMC article. Review. - Epigenetic Regulation of the Human Papillomavirus Life Cycle.
Mac M, Moody CA. Mac M, et al. Pathogens. 2020 Jun 18;9(6):483. doi: 10.3390/pathogens9060483. Pathogens. 2020. PMID: 32570816 Free PMC article. Review.
References
- Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006;580:6233–6241. - PubMed
- Bannasch D, Madge B, Schwab M. Functional interaction of Yaf2 with the central region of MycN. Oncogene. 2001;20:5913–5919. - PubMed
- Bejarano F, Gonzalez I, Vidal M, Busturia A. The Drosophila RYBP gene functions as a Polycomb-dependent transcriptional repressor. Mech Dev. 2005;122:1118–1129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases