Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods - PubMed (original) (raw)
Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods
Priit Adler et al. Genome Biol. 2009.
Abstract
We present a web resource MEM (Multi-Experiment Matrix) for gene expression similarity searches across many datasets. MEM features large collections of microarray datasets and utilizes rank aggregation to merge information from different datasets into a single global ordering with simultaneous statistical significance estimation. Unique features of MEM include automatic detection, characterization and visualization of datasets that includes the strongest coexpression patterns. MEM is freely available at http://biit.cs.ut.ee/mem/.
Figures
Figure 1
MEM user interface and results for the transcription factor NANOG. The top of the page contains controls for the query: gene input field, dataset selection and advanced options. Bottom of the page shows the results of the query. The genes, which are displayed as rows, are ordered by multi-experiment similarity to the query gene. Additionally, the single experiment similarity ranks are displayed as a matrix of colored squares, where red and blue denote small and large ranks, respectively. The larger squares indicate the ranks that contributed to the final _P_-value. Each element corresponds to a experiment and the columns are clustered. Hovering over the results brings up context specific information: (a) word cloud that characterizes the corresponding experiments; (b) single dataset annotations; (c) gene names with short descriptions. The row of links above the results facilitates the further analysis of results. For example, the user can visualize the expression of selected datasets (marked with green ticks) as a heat map (d).
Figure 2
NANOG targets among first 50 MEM results. MEM query with transcription factor NANOG retrieves more of its targets among top 50 genes, than queries on any one dataset individually. Each point represents the overlap between NANOG targets and top 50 query results in one of the 487 datasets. The datasets are sorted by variation and the ones that pass standard deviation filter are highlighted. Most of the datasets that retrieve high number of NANOG targets pass the filter, which shows the specificity of the filter.
Figure 3
Functional descriptions of the modules found in the mouse coexpression network constructed with MEM. Annotations of the six largest modules are shown in (a). Two smaller modules are shown in the Figure, along with their functional annotations in (b) and (c).
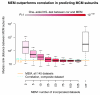
Figure 4
Increasing the number of datasets for MEM queries improves prediction of Mini Chromosome Maintenance (MCM) subunits. As additional datasets are incorporated for MEM analysis, MCM complex subunits show more consistent expression patterns as measured by median distance between subunits in MEM ranked lists of most correlated genes (decreasing bar height). According to one-sided Kolmogorov-Smirnov tests, MEM analysis with different numbers of datasets (left bars) significantly outperforms correlation (rightmost bar). In addition, MEM analysis for all the 145 selected datasets gives improved results compared to plain correlation across the concatenated dataset (light blue and orange lines).
Similar articles
- DRAGON View: information visualization for annotated microarray data.
Bouton CM, Pevsner J. Bouton CM, et al. Bioinformatics. 2002 Feb;18(2):323-4. doi: 10.1093/bioinformatics/18.2.323. Bioinformatics. 2002. PMID: 11847082 - PubMatrix: a tool for multiplex literature mining.
Becker KG, Hosack DA, Dennis G Jr, Lempicki RA, Bright TJ, Cheadle C, Engel J. Becker KG, et al. BMC Bioinformatics. 2003 Dec 10;4:61. doi: 10.1186/1471-2105-4-61. BMC Bioinformatics. 2003. PMID: 14667255 Free PMC article. - MILANO--custom annotation of microarray results using automatic literature searches.
Rubinstein R, Simon I. Rubinstein R, et al. BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article. - EXPath: a database of comparative expression analysis inferring metabolic pathways for plants.
Chien CH, Chow CN, Wu NY, Chiang-Hsieh YF, Hou PF, Chang WC. Chien CH, et al. BMC Genomics. 2015;16 Suppl 2(Suppl 2):S6. doi: 10.1186/1471-2164-16-S2-S6. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25708775 Free PMC article. - Bioinformatics analysis of microarray data.
Zhang Y, Szustakowski J, Schinke M. Zhang Y, et al. Methods Mol Biol. 2009;573:259-84. doi: 10.1007/978-1-60761-247-6_15. Methods Mol Biol. 2009. PMID: 19763933 Review.
Cited by
- Prediction of Tumor Microenvironment Characteristics and Treatment Response in Lung Squamous Cell Carcinoma by Pseudogene OR7E47P-related Immune Genes.
Zhao YQ, Zhang HH, Wu J, Li L, Li J, Zhong H, Jin Y, Lei TY, Zhao XY, Xu B, Song QB, He J. Zhao YQ, et al. Curr Med Sci. 2023 Dec;43(6):1133-1150. doi: 10.1007/s11596-023-2798-2. Epub 2023 Nov 28. Curr Med Sci. 2023. PMID: 38015361 - An insight into the potential role of LINC00968 in luminal breast cancer: Case-control study and bioinformatics analysis.
Arabpour M, Mehrpour Layeghi S, Majidzadeh-A K, Tavakkoly Bazzaz J, Mamivand A, Naghizadeh MM, Shakoori A. Arabpour M, et al. Biochem Biophys Rep. 2023 Aug 21;35:101531. doi: 10.1016/j.bbrep.2023.101531. eCollection 2023 Sep. Biochem Biophys Rep. 2023. PMID: 37654678 Free PMC article. - Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA.
Yang S, Ji J, Wang M, Nie J, Wang S. Yang S, et al. Biomolecules. 2023 Feb 6;13(2):306. doi: 10.3390/biom13020306. Biomolecules. 2023. PMID: 36830675 Free PMC article. - COVID-19 and New-Onset Psychosis: A Comprehensive Review.
Moccia L, Kotzalidis GD, Bartolucci G, Ruggiero S, Monti L, Biscosi M, Terenzi B, Ferrara OM, Mazza M, Di Nicola M, Janiri D, Simonetti A, Caroppo E, Janiri L, Sani G. Moccia L, et al. J Pers Med. 2023 Jan 2;13(1):104. doi: 10.3390/jpm13010104. J Pers Med. 2023. PMID: 36675765 Free PMC article. Review. - Gene co-expression analyses of health(span) across multiple species.
Möller S, Saul N, Projahn E, Barrantes I, Gézsi A, Walter M, Antal P, Fuellen G. Möller S, et al. NAR Genom Bioinform. 2022 Nov 29;4(4):lqac083. doi: 10.1093/nargab/lqac083. eCollection 2022 Dec. NAR Genom Bioinform. 2022. PMID: 36458022 Free PMC article.
References
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. - DOI - PubMed
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources