A multipathway phosphoproteomic signaling network model of idiosyncratic drug- and inflammatory cytokine-induced toxicity in human hepatocytes - PubMed (original) (raw)
A multipathway phosphoproteomic signaling network model of idiosyncratic drug- and inflammatory cytokine-induced toxicity in human hepatocytes
Benjamin D Cosgrove et al. Annu Int Conf IEEE Eng Med Biol Soc. 2009.
Abstract
Idiosyncratic drug hepatotoxicity is a hepatotoxicity subset that occurs in a very small fraction of human patients, is poorly predicted by standard preclinical models and in clinical trials, and frequently leads to postapproval drug failure. Animal models utilizing bacterial LPS co-administration to induce an inflammatory background and hepatocyte cell culture models utilizing cytokine mix cotreatment have successfully reproduced idiosyncratic hepatotoxicity signatures for certain drugs, but the hepatocyte signaling mechanisms governing these drug-cytokine toxicity synergizes are largely unclear. Here, we summarize our efforts to computationally model the signaling mechanisms regulating inflammatory cytokine-associated idiosyncratic drug hepatotoxicity. We collected a "cue-signal-response" (CSR) data compendium in cultured primary human hepatocytes treated with many combinations of idiosyncratic hepatotoxic drugs and inflammatory cytokine mixes ("cues") and subjected this compendium to orthogonal partial-least squares regression (OPLSR) to computationally relate the measured intracellular phosphoprotein signals and hepatocellular death responses. This OPLSR model suggested that hepatocytes specify their cell death responses to toxic drug/cytokine conditions by integrating signals from four key pathways - Akt, p70 S6K, ERK, and p38. An OPLSR model focused on data from these four signaling pathways demonstrated accurate predictions of idiosyncratic drug- and cytokine-induced hepatotoxicities in a second human hepatocyte donor, suggesting that hepatocytes from different individuals have shared network control mechanisms governing toxicity responses to diverse combinations of idiosyncratic hepatotoxicants and inflammatory cytokines.
Figures
Fig. 1
OPLSR modeling demonstrates accurate predictions of drug- and cytokine-induced hepatotoxicity across human hepatocyte donors. Phosphoprotein signaling and response data from two human hepatocyte donors [10]. An OPLSR model was trained on the 66-condition (only a subset of 6 conditions shown here), 6-phosphoprotein CSR data compendium from donor #1. This OPLSR model generated quantitatively accurate predictions of cell death responses in donor #1 and donor #2, even though donor-specific signaling network activation profiles and cell death responses were observed under the same drug/cytokine treatments. Compare clarithromycin (CLA) ± cytokine mix across the two donors. The predictive accuracy of this OPLSR model suggests that a common-effector processing mechanism (f(x) = y) encompassing the integration of the survival and stress signaling network (x) yields quantitatively concerted cell death responses (y) to toxic drug/cytokine conditions exists and is shared between hepatocytes from different human donors. Data and schematic reproduced from [10].
Fig. 2
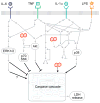
Inflammatory cytokine-associated idiosyncratic drug hepatotoxicity as a “network toxicity”. The multipathway modeling approach presented here suggests that an integration of multiple intracellular signaling pathway -- namely the MEK–ERK, mTOR–p70 S6K, Akt, and p38–HSP27 pathways -- activities is necessary for hepatocytes to specify death responses to hepatotoxic drug/cytokine co-treatment conditions. This provides motivation of the network-level consideration of multiple survival, stress, and apoptosis signaling pathways in evaluating the hepatotoxicity mechanisms of context-dependent hepatotoxic drugs. Schematic reproduced from [10].
Similar articles
- Cytokine-associated drug toxicity in human hepatocytes is associated with signaling network dysregulation.
Cosgrove BD, Alexopoulos LG, Hang TC, Hendriks BS, Sorger PK, Griffith LG, Lauffenburger DA. Cosgrove BD, et al. Mol Biosyst. 2010 Jul;6(7):1195-206. doi: 10.1039/b926287c. Epub 2010 Apr 1. Mol Biosyst. 2010. PMID: 20361094 Free PMC article. - Synergistic drug-cytokine induction of hepatocellular death as an in vitro approach for the study of inflammation-associated idiosyncratic drug hepatotoxicity.
Cosgrove BD, King BM, Hasan MA, Alexopoulos LG, Farazi PA, Hendriks BS, Griffith LG, Sorger PK, Tidor B, Xu JJ, Lauffenburger DA. Cosgrove BD, et al. Toxicol Appl Pharmacol. 2009 Jun 15;237(3):317-30. doi: 10.1016/j.taap.2009.04.002. Epub 2009 Apr 9. Toxicol Appl Pharmacol. 2009. PMID: 19362101 Free PMC article. - Omics-based identification of the combined effects of idiosyncratic drugs and inflammatory cytokines on the development of drug-induced liver injury.
Jiang J, Mathijs K, Timmermans L, Claessen SM, Hecka A, Weusten J, Peters R, van Delft JH, Kleinjans JCS, Jennen DGJ, de Kok TM. Jiang J, et al. Toxicol Appl Pharmacol. 2017 Oct 1;332:100-108. doi: 10.1016/j.taap.2017.07.014. Epub 2017 Jul 18. Toxicol Appl Pharmacol. 2017. PMID: 28733206 - Metabolic, idiosyncratic toxicity of drugs: overview of the hepatic toxicity induced by the anxiolytic, panadiplon.
Ulrich RG, Bacon JA, Brass EP, Cramer CT, Petrella DK, Sun EL. Ulrich RG, et al. Chem Biol Interact. 2001 May 16;134(3):251-70. doi: 10.1016/s0009-2797(01)00161-2. Chem Biol Interact. 2001. PMID: 11336974 Review. - Stem cell-derived models to improve mechanistic understanding and prediction of human drug-induced liver injury.
Goldring C, Antoine DJ, Bonner F, Crozier J, Denning C, Fontana RJ, Hanley NA, Hay DC, Ingelman-Sundberg M, Juhila S, Kitteringham N, Silva-Lima B, Norris A, Pridgeon C, Ross JA, Young RS, Tagle D, Tornesi B, van de Water B, Weaver RJ, Zhang F, Park BK. Goldring C, et al. Hepatology. 2017 Feb;65(2):710-721. doi: 10.1002/hep.28886. Epub 2016 Nov 30. Hepatology. 2017. PMID: 27775817 Free PMC article. Review.
Cited by
- Multiscale computational models of complex biological systems.
Walpole J, Papin JA, Peirce SM. Walpole J, et al. Annu Rev Biomed Eng. 2013;15:137-54. doi: 10.1146/annurev-bioeng-071811-150104. Epub 2013 Apr 29. Annu Rev Biomed Eng. 2013. PMID: 23642247 Free PMC article. Review. - Combined logical and data-driven models for linking signalling pathways to cellular response.
Melas IN, Mitsos A, Messinis DE, Weiss TS, Alexopoulos LG. Melas IN, et al. BMC Syst Biol. 2011 Jul 5;5:107. doi: 10.1186/1752-0509-5-107. BMC Syst Biol. 2011. PMID: 21729292 Free PMC article. - Using partial least squares regression to analyze cellular response data.
Kreeger PK. Kreeger PK. Sci Signal. 2013 Apr 16;6(271):tr7. doi: 10.1126/scisignal.2003849. Sci Signal. 2013. PMID: 23592846 Free PMC article. - Mechanistic Understanding of Idiosyncratic Drug-Induced Hepatotoxicity Using Co-Cultures of Hepatocytes and Macrophages.
Villanueva-Badenas E, Donato MT, Tolosa L. Villanueva-Badenas E, et al. Antioxidants (Basel). 2023 Jun 21;12(7):1315. doi: 10.3390/antiox12071315. Antioxidants (Basel). 2023. PMID: 37507855 Free PMC article. - Role of systems pharmacology in understanding drug adverse events.
Berger SI, Iyengar R. Berger SI, et al. Wiley Interdiscip Rev Syst Biol Med. 2011 Mar-Apr;3(2):129-35. doi: 10.1002/wsbm.114. Epub 2010 Aug 27. Wiley Interdiscip Rev Syst Biol Med. 2011. PMID: 20803507 Free PMC article. Review.
References
- Kaplowitz N. Drug Safety. 2001;24(7):483–490. - PubMed
- Lee WM. N Eng J Med. 2003;349(5):474–485. - PubMed
- Kaplowitz N. Nat Rev Drug Discov. 2005;4(6):489–499. - PubMed
- Ganey GE, Luyendyk JP, Maddox JF, Roth RA. Chem Biol Interact. 2004;150(1):35–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM068762/GM/NIGMS NIH HHS/United States
- U19 ES011399/ES/NIEHS NIH HHS/United States
- U19 ES 011399/ES/NIEHS NIH HHS/United States
- P50 GM 68762/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous