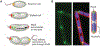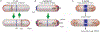Why and how bacteria localize proteins - PubMed (original) (raw)
Review
Why and how bacteria localize proteins
L Shapiro et al. Science. 2009.
Abstract
Despite their small size, bacteria have a remarkably intricate internal organization. Bacteria deploy proteins and protein complexes to particular locations and do so in a dynamic manner in lockstep with the organized deployment of their chromosome. The dynamic subcellular localization of protein complexes is an integral feature of regulatory processes of bacterial cells.
Figures
Fig. 1.
Localization patterns. (A) An anterior-posterior cellular localization axis is exhibited by the Caulobacter histidine kinases PleC (red) and DivJ (green) that dynamically and selectively localize to specific cell poles. The ZapA cell division protein (blue) localizes to the FtsZ ring. (B) In these same cells, a dorsal-ventral localization axis is exhibited by crescentin (cres) intermediate filaments that localize along the inner concave side of the crescent-shaped cell and are responsible for this distinctive cell shape (10); in contrast, the chemotaxis sensor array localizes at the convex outer side of the crescent near the cell pole (5). DAPI, 4′,6-diamidino-2-phenylindole.
Fig. 2.
Chromosome attachment to the cell pole. (A) The Caulobacter parS centromere bound to the ParB partition protein is attached to the cell pole by interaction with the polar PopZ polymeric network (14, 15). The initiation of replication triggers the assembly of PopZ at the opposite pole, where it captures the duplicated copy of parS/ParB. The diagram shows PopZ (red), ParB (green), PopZ + ParB (yellow), and chromosomes depicted as rings (dark green). (B) Sporulating cells of B. subtilis anchor chromosomes to the cell poles via the sporulation protein RacA, which binds to sites near the replication origin and to DivIVA at the cell poles (16, 17). The images show RacA tagged with green fluorescent protein (green), the nucleoids (blue), and the cell membrane (red).
Fig. 3.
Cell topology and inhibitor gradients control place and time of cell division. Repression of FtsZ polymerization by polar localized proteins that exhibit a minimum of the repressor at midcell restricts the site of division ring assembly in both E. coli and Caulobacter. However, _E. coli_’s strategy depends on oscillation of the MinCD repressor from pole to pole (A), whereas Caulobacter establishes a gradient of the MipZ repressor with the highest concentration at the cell poles (B). In S. pombe, the Pom1 repressor is localized to the poles (C). In small cells, the gradient of Pom1 extending from the poles overlaps at Cdr2 located at midcell and represses its activity. Consequently, the Cdk1 pathway is blocked, preventing entry into mitosis. As the cell grows, the midcell repressor concentration diminishes until Cdr2 (and thus the Cdk1 pathway) is no longer repressed and entry into mitosis is facilitated.
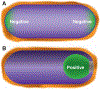
Fig. 4.
Geometric cues for protein localization. (A) Schematic depiction of the hemispherical poles of the cell. The more extreme negative curvature at the inside surface of the poles (relative to the inside surface of the lateral walls of the cell) could be a geometric cue for proteins, such as the cell division protein DivIVA, that localize to the poles. (B) The process of engulfment during spore formation in B. subtilis in which the membrane of the large mother cell (on the left) migrates around to surround and eventually pinch off the nascent spore. The positive curvature of the surface of the engulfment membrane from within the mother cell is a geometric cue for the sporulation protein SpoVM. The green color indicates the localization of DivIVA in (A) and SpoVM in (B) to regions of negative and positive curvature, respectively.
Similar articles
- Proteins on the move: dynamic protein localization in prokaryotes.
Jensen RB, Shapiro L. Jensen RB, et al. Trends Cell Biol. 2000 Nov;10(11):483-8. doi: 10.1016/s0962-8924(00)01840-7. Trends Cell Biol. 2000. PMID: 11050420 Review. - Generating and exploiting polarity in bacteria.
Shapiro L, McAdams HH, Losick R. Shapiro L, et al. Science. 2002 Dec 6;298(5600):1942-6. doi: 10.1126/science.1072163. Science. 2002. PMID: 12471245 Review. - Protein subcellular localization in bacteria.
Rudner DZ, Losick R. Rudner DZ, et al. Cold Spring Harb Perspect Biol. 2010 Apr;2(4):a000307. doi: 10.1101/cshperspect.a000307. Epub 2010 Mar 3. Cold Spring Harb Perspect Biol. 2010. PMID: 20452938 Free PMC article. Review. - Differentiation of the bacterial cell division site.
Cook WR, de Boer PA, Rothfield LI. Cook WR, et al. Int Rev Cytol. 1989;118:1-31. doi: 10.1016/s0074-7696(08)60871-2. Int Rev Cytol. 1989. PMID: 2691424 Review. No abstract available. - Subcellular Organization: A Critical Feature of Bacterial Cell Replication.
Surovtsev IV, Jacobs-Wagner C. Surovtsev IV, et al. Cell. 2018 Mar 8;172(6):1271-1293. doi: 10.1016/j.cell.2018.01.014. Cell. 2018. PMID: 29522747 Free PMC article. Review.
Cited by
- Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect.
Coquel AS, Jacob JP, Primet M, Demarez A, Dimiccoli M, Julou T, Moisan L, Lindner AB, Berry H. Coquel AS, et al. PLoS Comput Biol. 2013 Apr;9(4):e1003038. doi: 10.1371/journal.pcbi.1003038. Epub 2013 Apr 25. PLoS Comput Biol. 2013. PMID: 23633942 Free PMC article. - Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells.
Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Bakshi S, et al. Mol Microbiol. 2012 Jul;85(1):21-38. doi: 10.1111/j.1365-2958.2012.08081.x. Epub 2012 May 24. Mol Microbiol. 2012. PMID: 22624875 Free PMC article. - Life in a three-dimensional grid.
Shapiro L. Shapiro L. J Biol Chem. 2012 Nov 2;287(45):38289-94. doi: 10.1074/jbc.X112.422337. Epub 2012 Sep 24. J Biol Chem. 2012. PMID: 23007401 Free PMC article. - GerM is required to assemble the basal platform of the SpoIIIA-SpoIIQ transenvelope complex during sporulation in Bacillus subtilis.
Rodrigues CD, Ramírez-Guadiana FH, Meeske AJ, Wang X, Rudner DZ. Rodrigues CD, et al. Mol Microbiol. 2016 Oct;102(2):260-273. doi: 10.1111/mmi.13457. Epub 2016 Jul 22. Mol Microbiol. 2016. PMID: 27381174 Free PMC article. - Poles apart: prokaryotic polar organelles and their spatial regulation.
Kirkpatrick CL, Viollier PH. Kirkpatrick CL, et al. Cold Spring Harb Perspect Biol. 2011 Mar 1;3(3):a006809. doi: 10.1101/cshperspect.a006809. Cold Spring Harb Perspect Biol. 2011. PMID: 21084387 Free PMC article. Review.
References
- Alley MR, Maddock JR, Shapiro L, Genes Dev. 6, 825 (1992). - PubMed
- Maddock JR, Shapiro L, Science 259, 1717 (1993). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM07311/GM/NIGMS NIH HHS/United States
- R01 GM018568/GM/NIGMS NIH HHS/United States
- R24 GM073011/GM/NIGMS NIH HHS/United States
- T32 GM007311/GM/NIGMS NIH HHS/United States
- GM51426/GM/NIGMS NIH HHS/United States
- R01 GM051426/GM/NIGMS NIH HHS/United States
- R01 GM032506/GM/NIGMS NIH HHS/United States
- GM325062/GM/NIGMS NIH HHS/United States
- GM18568/GM/NIGMS NIH HHS/United States
- R37 GM018568/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources