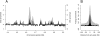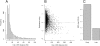Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading - PubMed (original) (raw)
Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading
Heather K MacAlpine et al. Genome Res. 2010 Feb.
Abstract
The origin recognition complex (ORC) is an essential DNA replication initiation factor conserved in all eukaryotes. In Saccharomyces cerevisiae, ORC binds to specific DNA elements; however, in higher eukaryotes, ORC exhibits little sequence specificity in vitro or in vivo. We investigated the genome-wide distribution of ORC in Drosophila and found that ORC localizes to specific chromosomal locations in the absence of any discernible simple motif. Although no clear sequence motif emerged, we were able to use machine learning approaches to accurately discriminate between ORC-associated sequences and ORC-free sequences based solely on primary sequence. The complex sequence features that define ORC binding sites are highly correlated with nucleosome positioning signals and likely represent a preferred nucleosomal landscape for ORC association. Open chromatin appears to be the underlying feature that is deterministic for ORC binding. ORC-associated sequences are enriched for the histone variant, H3.3, often at transcription start sites, and depleted for bulk nucleosomes. The density of ORC binding along the chromosome is reflected in the time at which a sequence replicates, with early replicating sequences having a high density of ORC binding. Finally, we found a high concordance between sites of ORC binding and cohesin loading, suggesting that, in addition to DNA replication, ORC may be required for the loading of cohesin on DNA in Drosophila.
Figures
Figure 1.
ORC localizes to specific chromosomal locations. (A) A subset of ORC binding sites map to early origins of replication. (Black) MA2C-normalized log2 ratios of ORC2 enrichment from chromatin immunoprecipitations are plotted as a function of chromosomal position for a 600-kb window of chromosome 3R. (Gray) Early origins were identified by immunoprecipitation of BrdU-containing replication intermediates during an HU arrest. (B) ORC is enriched at early origins of replication. The summits of 630 HU-resistant early origins were determined, and the average BrdU (gray) or ORC (black) enrichment is plotted relative to the distance from the early origin summit.
Figure 2.
Chromosomal density of ORC is a determinant of replication timing. (A) A histogram of inter-ORC distance throughout the genome. The median distance between ORC binding sites is 11 kb. (B) Time of replication is significantly delayed for sequences between distant ORC binding sites. The relative time of replication for the sequences equidistant from two ORC binding sites was plotted against the distance between the two ORC binding sites. (C) Early-replicating regions of the chromosome have an increased density of ORC binding. ORC binding sites per 100 kb were determined for the earliest and latest quartile of replication timing domains.
Figure 3.
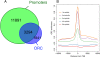
ORC associates with promoters of active genes. (A) Venn diagram depicting the overlap between ORC-associated sequences and annotated transcription start sites. (B) ORC localizes to the promoter regions of actively transcribed genes. The mRNA expression levels for approximately 15,000 Drosophila transcripts were determined by Affymetrix expression analysis. The mRNA expression levels for each gene were binned into five equal quintiles (Supplemental Fig. 2). The average ORC enrichment for each quintile of expression is plotted relative to the transcription start site.
Figure 4.
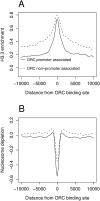
ORC localizes to open dynamic chromatin. (A) ORC binding sites are enriched for the histone variant H3.3. The average enrichment of H3.3 was plotted relative to the position of all ORC binding sites. The ORC binding sites were classified as either proximal or distal to annotated transcription start sites. Proximal ORC binding sites directly overlap with annotated promoters. (B) ORC binding sites are depleted for nucleosomes. The average depletion of nucleosomes was plotted relative to the position of promoter-proximal or -distal ORC binding sites.
Figure 5.
Cohesin loading occurs at ORC binding sites independent of pre-RC assembly. (A) ORC2 and SMC1 binding sites colocalize. ORC2 and SMC1 binding are indicated as blue density tracks for a 300-kb region of chromosome 2L. (B) ORC appears to nucleate cohesin binding. The average enrichments for ORC2 and SMC1 are plotted relative to the peak of all SMC1 binding sites in the Drosophila genome. (C) Cohesin loading is independent of pre-RC formation. Chromatin-associated MCM2-7, ORC, acetylated H4, and cohesin subunits at various points in the cell cycle (Supplemental Fig. 4).
Figure 6.
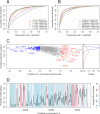
Sequence elements can discriminate between ORC-associated and ORC-free sequences. (A) ROC curves for SVM classifiers trained using as features the frequencies of _k_-mers for k = 1 to K, for increasing values of K. (B) ROC curves for SVM classifiers using subsets of features of increasing size. For each size of the feature set, we selected features with the highest absolute Pearson correlation with the class value (+1 or −1). All ROC curves were computed on the test set. (C) Correlation between _k_-mer features and class value, versus the average nucleosome occupancy over _k_-mers. The blue and red dots in the left plot correspond to the 500 features used in our analysis. The remaining features are shown in gray. The right plot shows the distributions of average nucleosome occupancy for the features with very high (red curve) and very low (blue curve) correlation coefficients. The average nucleosome occupancy values were computed from high-resolution in vitro nucleosome positioning data (Kaplan et al. 2009). (D) Predicted ORC binding for an arbitrary region on chromosome 3L. The background shows the average ORC2 ChIP enrichment; (blue) low enrichment; (pink) high enrichment. (Black curve) The posterior probability of ORC binding is in good agreement with the average ORC2 enrichment.
Similar articles
- [Ability of Su(Hw) to create a platform for ORC binding does not depend on the type of surrounding chromatin].
Mazina MIu, Vorob'eva NE, Krasnov AN. Mazina MIu, et al. Tsitologiia. 2013;55(4):218-24. Tsitologiia. 2013. PMID: 23875451 Russian. - Forced binding of the origin of replication complex to chromosomal sites in Drosophila S2 cells creates an origin of replication.
Crevel G, Cotterill S. Crevel G, et al. J Cell Sci. 2012 Feb 15;125(Pt 4):965-72. doi: 10.1242/jcs.094409. Epub 2012 Mar 15. J Cell Sci. 2012. PMID: 22421364 - Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome.
Vorobyeva NE, Mazina MU, Golovnin AK, Kopytova DV, Gurskiy DY, Nabirochkina EN, Georgieva SG, Georgiev PG, Krasnov AN. Vorobyeva NE, et al. Nucleic Acids Res. 2013 Jun;41(11):5717-30. doi: 10.1093/nar/gkt297. Epub 2013 Apr 22. Nucleic Acids Res. 2013. PMID: 23609538 Free PMC article. - Changing the DNA landscape: putting a SPN on chromatin.
Formosa T. Formosa T. Curr Top Microbiol Immunol. 2003;274:171-201. doi: 10.1007/978-3-642-55747-7_7. Curr Top Microbiol Immunol. 2003. PMID: 12596908 Review. - Multiple functions of the origin recognition complex.
Chesnokov IN. Chesnokov IN. Int Rev Cytol. 2007;256:69-109. doi: 10.1016/S0074-7696(07)56003-1. Int Rev Cytol. 2007. PMID: 17241905 Review.
Cited by
- Open chromatin structures regulate the efficiencies of pre-RC formation and replication initiation in Epstein-Barr virus.
Papior P, Arteaga-Salas JM, Günther T, Grundhoff A, Schepers A. Papior P, et al. J Cell Biol. 2012 Aug 20;198(4):509-28. doi: 10.1083/jcb.201109105. Epub 2012 Aug 13. J Cell Biol. 2012. PMID: 22891264 Free PMC article. - How the cell cycle impacts chromatin architecture and influences cell fate.
Ma Y, Kanakousaki K, Buttitta L. Ma Y, et al. Front Genet. 2015 Feb 3;6:19. doi: 10.3389/fgene.2015.00019. eCollection 2015. Front Genet. 2015. PMID: 25691891 Free PMC article. - Genome maintenance in the context of 4D chromatin condensation.
Yu S, Yang F, Shen WH. Yu S, et al. Cell Mol Life Sci. 2016 Aug;73(16):3137-50. doi: 10.1007/s00018-016-2221-2. Epub 2016 Apr 20. Cell Mol Life Sci. 2016. PMID: 27098512 Free PMC article. Review. - Chromatin landscape: methylation beyond transcription.
Black JC, Whetstine JR. Black JC, et al. Epigenetics. 2011 Jan;6(1):9-15. doi: 10.4161/epi.6.1.13331. Epub 2011 Jan 1. Epigenetics. 2011. PMID: 20855937 Free PMC article. Review. - The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks.
van Schie JJM, de Lange J. van Schie JJM, et al. Cells. 2021 Dec 8;10(12):3455. doi: 10.3390/cells10123455. Cells. 2021. PMID: 34943967 Free PMC article. Review.
References
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. - PubMed
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM081883/GM/NIGMS NIH HHS/United States
- U01 HG004279/HG/NHGRI NIH HHS/United States
- HG004279/HG/NHGRI NIH HHS/United States
- P50-GM081883-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases