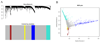Gene networks and microRNAs implicated in aggressive prostate cancer - PubMed (original) (raw)
Gene networks and microRNAs implicated in aggressive prostate cancer
Liang Wang et al. Cancer Res. 2009.
Abstract
Prostate cancer, a complex disease, can be relatively harmless or extremely aggressive. To identify candidate genes involved in causal pathways of aggressive prostate cancer, we implemented a systems biology approach by combining differential expression analysis and coexpression network analysis to evaluate transcriptional profiles using lymphoblastoid cell lines from 62 prostate cancer patients with aggressive phenotype (Gleason grade >or= 8) and 63 prostate cancer patients with nonaggressive phenotype (Gleason grade <or= 5). From 13,935 mRNA genes and 273 microRNAs (miRNA) tested, we identified significant differences in 1,100 mRNAs and 7 miRNAs with a false discovery rate (FDR) of <0.01. We also identified a coexpression module demonstrating significant association with the aggressive phenotype of prostate cancer (P = 3.67 x 10(-11)). The module of interest was characterized by overrepresentation of cell cycle-related genes (FDR = 3.50 x 10(-50)). From this module, we further defined 20 hub genes that were highly connected to other genes. Interestingly, 5 of the 7 differentially expressed miRNAs have been implicated in cell cycle regulation and 2 (miR-145 and miR-331-3p) are predicted to target 3 of the 20 hub genes. Ectopic expression of these two miRNAs reduced expression of target hub genes and subsequently resulted in cell growth inhibition and apoptosis. These results suggest that cell cycle is likely to be a molecular pathway causing aggressive phenotype of prostate cancer. Further characterization of cell cycle-related genes (particularly, the hub genes) and miRNAs that regulate these hub genes could facilitate identification of candidate genes responsible for the aggressive phenotype and lead to a better understanding of prostate cancer etiology and progression.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures
Figure 1. Gene co-expression network analysis
A. Branches (gene modules) of highly correlated genes by average linkage hierarchical clustering of 3100 genes. The colored bars directly corresponded to the module (color) designation for the clusters of genes. Grey denoted genes that were not part of any module. The remaining colors were used for the four modules. B. Multi-dimensional scaling plot of the entire gene expression network. Each dot represented a gene, where the color corresponded to the gene module. The distance between each dot indicated their topological overlap.
Figure 2. Identification of clinical trait-related hub genes
A. Scatterplot between gene significance (absolute r) (_y_-axis) and scaled intramodular connectivity (K/Kmax). Each point corresponded to a gene in the turquoise module. The intramodular connectivity was significantly correlated with gene significance (r = 0.61, p = 7.1 × 10−19). B. Visualization of gene-gene interaction within turquoise module. The connections were drawn using VisANT tool (ref 22). The genes with at least one connection when weighted cutoff value >=0.16 were shown. Each node represented a gene. Red nodes were hub genes. Bigger nodes indicated more connections.
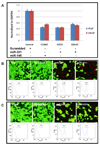
Figure 3. Biological effect of ectopic expression of miR-145 and miR-331 in prostate cancer cell lines
A. qRT-PCR was used to measure expression level of target genes using total RNA from nucleofected cells. Expression values were normalized to GAPDH. Expression levels of target genes were significantly reduced by ectopic expression of the two miRNAs. Cell viability was examined in VCaP cells (B) and LNCaP cells (C). Live and dead cells were stained in green and red, respectively. Percentage of apoptotic cell population measured by FACS was shown below each corresponding cell image.
Similar articles
- Identification of Potential miRNAs Biomarkers for High-Grade Prostate Cancer by Integrated Bioinformatics Analysis.
Foj L, Filella X. Foj L, et al. Pathol Oncol Res. 2019 Oct;25(4):1445-1456. doi: 10.1007/s12253-018-0508-3. Epub 2018 Oct 26. Pathol Oncol Res. 2019. PMID: 30367364 - Construction and analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal the key genes associated with prostate cancer.
Ye Y, Li SL, Wang SY. Ye Y, et al. PLoS One. 2018 Aug 23;13(8):e0198055. doi: 10.1371/journal.pone.0198055. eCollection 2018. PLoS One. 2018. PMID: 30138363 Free PMC article. - miRNA-mRNA correlation-network modules in human prostate cancer and the differences between primary and metastatic tumor subtypes.
Zhang W, Edwards A, Fan W, Flemington EK, Zhang K. Zhang W, et al. PLoS One. 2012;7(6):e40130. doi: 10.1371/journal.pone.0040130. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768240 Free PMC article. - MicroRNA-guided gene expression in prostate cancer: Literature and database overview.
Bryzgunova OE, Konoshenko MY, Laktionov PP. Bryzgunova OE, et al. J Gene Med. 2018 May;20(5):e3016. doi: 10.1002/jgm.3016. Epub 2018 Apr 30. J Gene Med. 2018. PMID: 29578262 Review. - Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer.
Coradduzza D, Cruciani S, Arru C, Garroni G, Pashchenko A, Jedea M, Zappavigna S, Caraglia M, Amler E, Carru C, Maioli M. Coradduzza D, et al. Int J Mol Sci. 2022 Jan 30;23(3):1626. doi: 10.3390/ijms23031626. Int J Mol Sci. 2022. PMID: 35163550 Free PMC article. Review.
Cited by
- Hsa_circ_0038646 promotes cell proliferation and migration in colorectal cancer via miR-331-3p/GRIK3.
Du H, He Z, Feng F, Chen D, Zhang L, Bai J, Wu H, Han E, Zhang J. Du H, et al. Oncol Lett. 2020 Jul;20(1):266-274. doi: 10.3892/ol.2020.11547. Epub 2020 Apr 21. Oncol Lett. 2020. PMID: 32565953 Free PMC article. - Integrated network analysis and logistic regression modeling identify stage-specific genes in Oral Squamous Cell Carcinoma.
Randhawa V, Acharya V. Randhawa V, et al. BMC Med Genomics. 2015 Jul 16;8:39. doi: 10.1186/s12920-015-0114-0. BMC Med Genomics. 2015. PMID: 26179909 Free PMC article. - Androgen receptor-mediated downregulation of microRNA-221 and -222 in castration-resistant prostate cancer.
Gui B, Hsieh CL, Kantoff PW, Kibel AS, Jia L. Gui B, et al. PLoS One. 2017 Sep 8;12(9):e0184166. doi: 10.1371/journal.pone.0184166. eCollection 2017. PLoS One. 2017. PMID: 28886115 Free PMC article. - Integrated Analysis of Three Publicly Available Gene Expression Profiles Identified Genes and Pathways Associated with Clear Cell Renal Cell Carcinoma.
Han Y, Wang L, Wang Y. Han Y, et al. Med Sci Monit. 2020 Jul 26;26:e919965. doi: 10.12659/MSM.919965. Med Sci Monit. 2020. PMID: 32712616 Free PMC article. - CASC11 promotes aggressiveness of prostate cancer cells through miR-145/IGF1R axis.
Capik O, Sanli F, Kurt A, Ceylan O, Suer I, Kaya M, Ittmann M, Karatas OF. Capik O, et al. Prostate Cancer Prostatic Dis. 2021 Sep;24(3):891-902. doi: 10.1038/s41391-021-00353-0. Epub 2021 Mar 22. Prostate Cancer Prostatic Dis. 2021. PMID: 33753875
References
Publication types
MeSH terms
Substances
Grants and funding
- R21 CA126786/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- P30 CA15083/CA/NCI NIH HHS/United States
- CA126786/CA/NCI NIH HHS/United States
- CA91956/CA/NCI NIH HHS/United States
- P50 CA091956/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases