Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase - PubMed (original) (raw)
. 2010 Feb 19;285(8):5792-801.
doi: 10.1074/jbc.M109.065516. Epub 2009 Dec 9.
Ricardo Garcia-Serres, Geneviève Blondin, Thierry Douki, Martin Clemancey, Jean-Marc Latour, Farhad Forouhar, Helen Neely, Gaetano T Montelione, John F Hunt, Etienne Mulliez, Marc Fontecave, Mohamed Atta
Affiliations
- PMID: 20007320
- PMCID: PMC2820805
- DOI: 10.1074/jbc.M109.065516
Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase
Simon Arragain et al. J Biol Chem. 2010.
Abstract
Post-translational modifications of ribosomal proteins are important for the accuracy of the decoding machinery. A recent in vivo study has shown that the rimO gene is involved in generation of the 3-methylthio derivative of residue Asp-89 in ribosomal protein S12 (Anton, B. P., Saleh, L., Benner, J. S., Raleigh, E. A., Kasif, S., and Roberts, R. J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1826-1831). This reaction is formally identical to that catalyzed by MiaB on the C2 of adenosine 37 near the anticodon of several tRNAs. We present spectroscopic evidence that Thermotoga maritima RimO, like MiaB, contains two [4Fe-4S] centers, one presumably bound to three invariant cysteines in the central radical S-adenosylmethionine (AdoMet) domain and the other to three invariant cysteines in the N-terminal UPF0004 domain. We demonstrate that holo-RimO can specifically methylthiolate the aspartate residue of a 20-mer peptide derived from S12, yielding a mixture of mono- and bismethylthio derivatives. Finally, we present the 2.0 A crystal structure of the central radical AdoMet and the C-terminal TRAM (tRNA methyltransferase 2 and MiaB) domains in apo-RimO. Although the core of the open triose-phosphate isomerase (TIM) barrel of the radical AdoMet domain was conserved, RimO showed differences in domain organization compared with other radical AdoMet enzymes. The unusually acidic TRAM domain, likely to bind the basic S12 protein, is located at the distal edge of the radical AdoMet domain. The basic S12 protein substrate is likely to bind RimO through interactions with both the TRAM domain and the concave surface of the incomplete TIM barrel. These biophysical results provide a foundation for understanding the mechanism of methylthioation by radical AdoMet enzymes in the MiaB/RimO family.
Figures
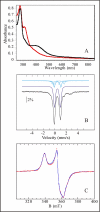
FIGURE 1.
Spectroscopic characterization of holo-RimO. A, UV-visible absorption spectra of oxidized (black) and reduced (red) forms of holo-RimO (8 μ
m
) in 50 m
m
NaCl, 50 m
m
Tris-HCl, pH 8. The absorbance at 314 nm is due to excess dithionite. B, Mössbauer spectrum of holo-RimO (288 μ
m
) in the same buffer. The experimental spectrum (hatched marks) was recorded at 4.2 K in a magnetic field of 500 mT applied perpendicular to the direction of the γ-beam. The theoretical spectra of the different species are shown as colored lines above the experimental spectrum. The dark blue line represents a typical [4Fe-4S]2+ cluster, and the light blue lines represent the three distinct sites of an atypical [4Fe-4S]2+ cluster composed of a delocalized FeIIFeIII pair (solid line) and two distinct Fe centers, one being more ferrous in character (hatched line) than the other (dotted line). The parameters of all these doublets and their relative absorption intensities are given under “Results.” Additionally, two quadrupole doublets (not shown) representing adventitiously bound FeII were used to simulate the spectrum. The composite spectrum is plotted as a solid black line overlaid with the experimental data. C, X-band EPR spectrum of reduced holo-RimO (144 μ
m
) in the same buffer. The dotted red line shows the g = 2 region of the X-band EPR spectrum recorded on a dithionite reduced sample (natural 57Fe abundance) at 20 K under nonsaturating conditions (0.63-milliwatt microwave power at 9.65 GHz frequency). The blue line shows a theoretical simulation with two S = ½ species in a 51:49 ratio using a Lorentzian profile. The parameters of species 1 are g1x = 1.902, g1y = 1.930, g1z = 2.030, w1x = 2.7 mT, w1y = 3.4 mT, and w1z = 2.2 mT, whereas those of species 2 are g2x = 1.879, g2y = 1.940, g2z = 2.042, w2x = 3.4 mT, w2y = 1.1 mT, and w2z = 3.1 mT (half-width at half-maximum).
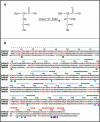
SCHEME 1.
A, the reaction catalyzed by RimO; “S_” represents the sulfur atom donor. B, sequence alignment of T. maritima RimO (TM1862), MiaB (TM0653), and TM0830 performed using T-COFFEE (41). TM0830 is a third paralog with equivalent domain organization also encoded in the genome of T. maritima. Identical residues in all three paralogs are shown in red. A structural schematic is shown above the alignment with α-helices and β-strands represented by rectangles and arrows, respectively, and backbone segments not observed in the crystal structure of apo_-T. maritima RimO are indicated by dotted lines. The UPF0004 domain is shown in green, the radical AdoMet domain in yellow and blue, and the TRAM domain in orange and red. (None of the UPF0004 domain was observed in the crystal structure.) The green circles below the alignment indicate the invariant cysteines, and the cyan circles indicate conserved residues making salt bridges in the interface between the radical AdoMet and TRAM domains. The magenta and purple circles indicate, respectively, invariant and conserved acidic residues among RimO orthologs (data not shown) on the surface of the TRAM domain proximal to the active site of the radical AdoMet domain.
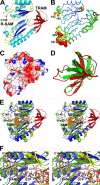
FIGURE 2.
Crystal structure of the radical AdoMet and TRAM domains in apo-RimO from T. maritima. A, schematic diagram with the radical _S_-adenosylmethionine (R-SAM) domain colored cyan, blue, and yellow and the TRAM domain colored red and orange. Invariant residue Cys-148, which probably ligates one [4Fe-4S] cluster in holo-RimO, is shown in ball-and-stick representation. B, an equivalent view of the structure with the color and thickness of the backbone worm representing backbone B-factors, which measure the degree of positional disorder in the crystal. The color ramp runs from blue for the lowest B-factors (10.7 Å2) to red for the highest (69.1 Å2). The C terminus is labeled Cter, and residue numbers are given for all other termini of the observed polypeptide segments. C, electrostatic surface potential of RimO, with blue and red representing acidic and basic regions, respectively, and fully saturated colors indicating a potential of ±14 kT at an ionic strength of 100 m
m
(24). D, structural superposition of the TRAM domains in RimO (red) and RumA (green) as aligned by DALI. E, stereopair showing structural superposition of the radical AdoMet domains in RimO (blue and red) and MoaA (green and yellow) as aligned by DALI. The AdoMet co-substrate (orange) and GTP substrate (wheat) in MoaA are shown in space-filling representations, and its [4Fe-4S] clusters and Cys-24 (equivalent to Cys-148 in RimO) are shown in ball-and-stick representations. The C-terminal α-helices in MoaA, which pack on the surface of the yellow β-sheets, have been omitted to improve clarity. F, stereopair showing a close-up view of the active site in the same structural superposition.
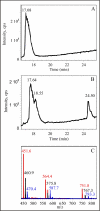
FIGURE 3.
HPLC-MS identification of enzymatic reaction products. A, total ion-current chromatogram for the 20-mer peptide without enzyme treatment. B, equivalent chromatogram after reaction of the peptide with holo-RimO. The peaks at 18.55 and 25.50 min correspond to monomethylthio and bismethylthio derivatives, respectively. C, mass spectra of the unmodified peptide (red), monomethylthio derivative (black), and bismethylthio derivative (blue) showing +5, +4, and +3 charge states.
Similar articles
- Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily.
Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ. Lee KH, et al. Biochemistry. 2009 Oct 27;48(42):10162-74. doi: 10.1021/bi900939w. Biochemistry. 2009. PMID: 19736993 Free PMC article. - RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli.
Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. Anton BP, et al. Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):1826-31. doi: 10.1073/pnas.0708608105. Epub 2008 Feb 5. Proc Natl Acad Sci U S A. 2008. PMID: 18252828 Free PMC article. - Stereochemical Course of the Reaction Catalyzed by RimO, a Radical SAM Methylthiotransferase.
Landgraf BJ, Booker SJ. Landgraf BJ, et al. J Am Chem Soc. 2016 Mar 9;138(9):2889-92. doi: 10.1021/jacs.5b11035. Epub 2016 Feb 25. J Am Chem Soc. 2016. PMID: 26871608 - Structural diversity in the AdoMet radical enzyme superfamily.
Dowling DP, Vey JL, Croft AK, Drennan CL. Dowling DP, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1178-95. doi: 10.1016/j.bbapap.2012.04.006. Epub 2012 Apr 28. Biochim Biophys Acta. 2012. PMID: 22579873 Free PMC article. Review. - SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes.
Grell TA, Goldman PJ, Drennan CL. Grell TA, et al. J Biol Chem. 2015 Feb 13;290(7):3964-71. doi: 10.1074/jbc.R114.581249. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477505 Free PMC article. Review.
Cited by
- Identification of an intermediate methyl carrier in the radical S-adenosylmethionine methylthiotransferases RimO and MiaB.
Landgraf BJ, Arcinas AJ, Lee KH, Booker SJ. Landgraf BJ, et al. J Am Chem Soc. 2013 Oct 16;135(41):15404-15416. doi: 10.1021/ja4048448. Epub 2013 Oct 3. J Am Chem Soc. 2013. PMID: 23991893 Free PMC article. - Mechanistic and functional versatility of radical SAM enzymes.
Booker SJ, Grove TL. Booker SJ, et al. F1000 Biol Rep. 2010 Jul 14;2:52. doi: 10.3410/B2-52. F1000 Biol Rep. 2010. PMID: 21152342 Free PMC article. - Biochemical Approaches to Probe the Role of the Auxiliary Iron-Sulfur Cluster of Lipoyl Synthase from Mycobacterium Tuberculosis.
Jeyachandran VR, Pendyala JV, McCarthy EL, Boal AK, Booker SJ. Jeyachandran VR, et al. Methods Mol Biol. 2021;2353:307-332. doi: 10.1007/978-1-0716-1605-5_16. Methods Mol Biol. 2021. PMID: 34292556 - Post-translational modification of ribosomally synthesized peptides by a radical SAM epimerase in Bacillus subtilis.
Benjdia A, Guillot A, Ruffié P, Leprince J, Berteau O. Benjdia A, et al. Nat Chem. 2017 Jul;9(7):698-707. doi: 10.1038/nchem.2714. Epub 2017 Feb 6. Nat Chem. 2017. PMID: 28644475 Free PMC article. - Structural insights into radical generation by the radical SAM superfamily.
Vey JL, Drennan CL. Vey JL, et al. Chem Rev. 2011 Apr 13;111(4):2487-506. doi: 10.1021/cr9002616. Epub 2011 Mar 3. Chem Rev. 2011. PMID: 21370834 Free PMC article. Review. No abstract available.
References
- Carroll A. J., Heazlewood J. L., Ito J., Millar A. H. (2008) Mol. Cell. Proteomics 7, 347–369 - PubMed
- Polevoda B., Sherman F. (2007) Mol. Microbiol. 65, 590–606 - PubMed
- Dong H., Kurland C. G. (1995) J. Mol. Biol. 248, 551–561 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous