Network analyses structure genetic diversity in independent genetic worlds - PubMed (original) (raw)
Network analyses structure genetic diversity in independent genetic worlds
Sébastien Halary et al. Proc Natl Acad Sci U S A. 2010.
Abstract
DNA flows between chromosomes and mobile elements, following rules that are poorly understood. This limited knowledge is partly explained by the limits of current approaches to study the structure and evolution of genetic diversity. Network analyses of 119,381 homologous DNA families, sampled from 111 cellular genomes and from 165,529 phage, plasmid, and environmental virome sequences, offer challenging insights. Our results support a disconnected yet highly structured network of genetic diversity, revealing the existence of multiple "genetic worlds." These divides define multiple isolated groups of DNA vehicles drawing on distinct gene pools. Mathematical studies of the centralities of these worlds' subnetworks demonstrate that plasmids, not viruses, were key vectors of genetic exchange between bacterial chromosomes, both recently and in the past. Furthermore, network methodology introduces new ways of quantifying current sampling of genetic diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Network of shared DNA families among cellular, plasmid, and phage genomes. (A) Global network in which each node represents a genome, either cellular (green for bacterial chromosome, yellow for eukaryotic chromosomes, and cyan for archaeal chromosome), plasmidic (purple), or phage (red). Two nodes are connected by an edge if they share homologous DNA (reciprocal best BLAST hit with a minimum of 1e-20 score, and 20% minimum identity). Edges are weighted by the number of shared DNA families. The layout was produced by Cytoscape, using an edge-weighted spring-embedded model, meaning that genomes sharing more DNA families are closer on the display. There are 3,207 nodes on that network. (B) Global network displaying connections between genomes (same color code) for a minimum of 95% identity. Imposing a minimum identity percentage on the definition of CHDs roughly filters for more recent sharing events. Bacterial clusters are indicated as follows: Burkholderia (1), Xanthomonas (2), Yersinia, (3) Streptococcus (4), Prochlorococcus/Synechocystis (5), Clostridium (6), Legionella (7), Rhodopseudomonas (8), and Helicobacter (9). There are 1,529 nodes on that network.
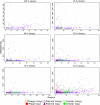
Fig. 2.
Betweenness of nodes as function of their degree for various identity threshold networks. Cellular chromosomes are displayed as green circles, plasmids as purple triangles, and phages as red squares. When the betweenness of a node is significantly higher than expected (P < 0.05), the corresponding symbol is filled, and it is empty otherwise. Although betweenness generally increases with degree, plasmids and cellular clearly show higher betweenness values, suggesting they play a central role in the sharing of DNA. Note that scale differs among plots. There are 171 nodes for a 100% identity threshold, 342 for a 95% identity threshold, 372 for a 90% identity threshold, 509 for a 80% identity threshold, 618 for a 70% identity threshold, and 1,029 for a 40% identity threshold.
Similar articles
- Plasmid diversity in neisseriae.
van Passel MW, van der Ende A, Bart A. van Passel MW, et al. Infect Immun. 2006 Aug;74(8):4892-9. doi: 10.1128/IAI.02087-05. Infect Immun. 2006. PMID: 16861678 Free PMC article. - Bipartite Network Analysis of the Archaeal Virosphere: Evolutionary Connections between Viruses and Capsidless Mobile Elements.
Iranzo J, Koonin EV, Prangishvili D, Krupovic M. Iranzo J, et al. J Virol. 2016 Nov 28;90(24):11043-11055. doi: 10.1128/JVI.01622-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681128 Free PMC article. - Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences.
Lanza VF, de Toro M, Garcillán-Barcia MP, Mora A, Blanco J, Coque TM, de la Cruz F. Lanza VF, et al. PLoS Genet. 2014 Dec 18;10(12):e1004766. doi: 10.1371/journal.pgen.1004766. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25522143 Free PMC article. - Ten years of bacterial genome sequencing: comparative-genomics-based discoveries.
Binnewies TT, Motro Y, Hallin PF, Lund O, Dunn D, La T, Hampson DJ, Bellgard M, Wassenaar TM, Ussery DW. Binnewies TT, et al. Funct Integr Genomics. 2006 Jul;6(3):165-85. doi: 10.1007/s10142-006-0027-2. Epub 2006 May 12. Funct Integr Genomics. 2006. PMID: 16773396 Review. - Pathogenomics of the Ralstonia solanacearum species complex.
Genin S, Denny TP. Genin S, et al. Annu Rev Phytopathol. 2012;50:67-89. doi: 10.1146/annurev-phyto-081211-173000. Epub 2012 May 1. Annu Rev Phytopathol. 2012. PMID: 22559068 Review.
Cited by
- Abiotic Gene Transfer: Rare or Rampant?
Kotnik T, Weaver JC. Kotnik T, et al. J Membr Biol. 2016 Oct;249(5):623-631. doi: 10.1007/s00232-016-9897-y. Epub 2016 Apr 11. J Membr Biol. 2016. PMID: 27067073 Free PMC article. Review. - "Every Gene Is Everywhere but the Environment Selects": Global Geolocalization of Gene Sharing in Environmental Samples through Network Analysis.
Fondi M, Karkman A, Tamminen MV, Bosi E, Virta M, Fani R, Alm E, McInerney JO. Fondi M, et al. Genome Biol Evol. 2016 May 13;8(5):1388-400. doi: 10.1093/gbe/evw077. Genome Biol Evol. 2016. PMID: 27190206 Free PMC article. - Cargo capacity of phages and plasmids and other factors influencing horizontal transfers of prokaryote transposable elements.
Leclercq S, Gilbert C, Cordaux R. Leclercq S, et al. Mob Genet Elements. 2012 Mar 1;2(2):115-118. doi: 10.4161/mge.20352. Mob Genet Elements. 2012. PMID: 22934247 Free PMC article. - Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements.
Humphrey S, Fillol-Salom A, Quiles-Puchalt N, Ibarra-Chávez R, Haag AF, Chen J, Penadés JR. Humphrey S, et al. Nat Commun. 2021 Nov 8;12(1):6509. doi: 10.1038/s41467-021-26004-5. Nat Commun. 2021. PMID: 34750368 Free PMC article. - Of woods and webs: possible alternatives to the tree of life for studying genomic fluidity in E. coli.
Beauregard-Racine J, Bicep C, Schliep K, Lopez P, Lapointe FJ, Bapteste E. Beauregard-Racine J, et al. Biol Direct. 2011 Jul 20;6:39; discussion 39. doi: 10.1186/1745-6150-6-39. Biol Direct. 2011. PMID: 21774799 Free PMC article.
References
- Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous