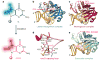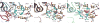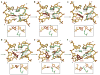Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme - PubMed (original) (raw)
Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme
Yan Qi et al. Nature. 2009.
Abstract
How living systems detect the presence of genotoxic damage embedded in a million-fold excess of undamaged DNA is an unresolved question in biology. Here we have captured and structurally elucidated a base-excision DNA repair enzyme, MutM, at the stage of initial encounter with a damaged nucleobase, 8-oxoguanine (oxoG), nested within a DNA duplex. Three structures of intrahelical oxoG-encounter complexes are compared with sequence-matched structures containing a normal G base in place of an oxoG lesion. Although the protein-DNA interfaces in the matched complexes differ by only two atoms-those that distinguish oxoG from G-their pronounced structural differences indicate that MutM can detect a lesion in DNA even at the earliest stages of encounter. All-atom computer simulations show the pathway by which encounter of the enzyme with the lesion causes extrusion from the DNA duplex, and they elucidate the critical free energy difference between oxoG and G along the extrusion pathway.
Figures
Figure 1. Generation and recognition of 8-oxoguanine
a, Structural comparison of G versus oxoG, with differences highlighted. b, Overall structure of a lesion-recognition complex, LRC3, with an extrahelical oxoG lesion bound in the enzyme active site. c, Sequence-matched interrogation complex, IC3 (ref. 14), with a fully intrahelical target G•C base-pair. d, Close-up view of the oxoG-capping loop (OCL) and its contacts to oxoG in LRC3. The light pink region represents the residues deleted in EC3–5. Residues with grey side chains were mutated to prolines in the ECs bearing OCL point mutations. e, Sequence-matched encounter complex, EC3, bearing a fully intrahelical target oxoG•C base-pair.
Figure 2. Helix-penetration by MutM residues
a, LRC3; b, IC3; c, EC3. Colour-coding is as in Fig. 1, except side chains of the key residues (M77, R112, F114 and E78) are shown in cyan.
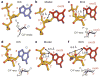
Figure 3. OxoG-dependent local DNA structure alterations at the site of the target base in EC3 and EC5
a and d, target G; c and f, target oxoG; b and e, models constructed by the addition of a carbonyl oxygen to the 8-position of the corresponding target G residue in a and d, respectively. The perspectives were chosen to facilitate examination of sugar puckers (diagrammed below a, c, d, and f). Steric clashes in the models are indicated by double-headed arrows.
Figure 4. Free energy profiles of nucleobase extrusion imply active participation of MutM
a, Free energies along the base extrusion pathways, as a function of the normalized arc length, Z, in b and Supplementary Fig. 17. Critical events associated with main energetic barriers are marked. b, Free energy landscape plots for extrusion of oxoG starting from EC4 (top) and G from IC4 (bottom). See Supplementary Fig. 16 for definitions of _q_ext and _q_rotat. The black dotted lines denote the minimal free energy paths for base extrusion, which define Z. The lowest free energy points in the intrahelical states are set to zero.
Figure 5. R112-catalysed oxoG extrusion
a –f, Snapshots from targeted molecular dynamics (TMD) simulations in the vicinity of the target base, showing helix invasion by R112 and concomitant extrusion of the target oxoG. The three DNA helix-invading MutM residues (M77, R112 and F114) are coloured in green. Nucleosides around the target oxoG•C pair are shown in gold. The inset figure of each panel is an orthogonal view. See the text for the progression of events a–f. Refer to the Supplementary Movies for the complete base extrusion trajectories of oxoG and G by MutM.
Similar articles
- DNA lesion recognition by the bacterial repair enzyme MutM.
Fromme JC, Verdine GL. Fromme JC, et al. J Biol Chem. 2003 Dec 19;278(51):51543-8. doi: 10.1074/jbc.M307768200. Epub 2003 Oct 1. J Biol Chem. 2003. PMID: 14525999 - Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
Sung RJ, Zhang M, Qi Y, Verdine GL. Sung RJ, et al. J Biol Chem. 2012 May 25;287(22):18044-54. doi: 10.1074/jbc.M111.313635. Epub 2012 Mar 30. J Biol Chem. 2012. PMID: 22465958 Free PMC article. - Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM.
Qi Y, Spong MC, Nam K, Karplus M, Verdine GL. Qi Y, et al. J Biol Chem. 2010 Jan 8;285(2):1468-78. doi: 10.1074/jbc.M109.069799. Epub 2009 Nov 4. J Biol Chem. 2010. PMID: 19889642 Free PMC article. - Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions.
Hazra TK, Hill JW, Izumi T, Mitra S. Hazra TK, et al. Prog Nucleic Acid Res Mol Biol. 2001;68:193-205. doi: 10.1016/s0079-6603(01)68100-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554297 Review. - DNA glycosylase recognition and catalysis.
Fromme JC, Banerjee A, Verdine GL. Fromme JC, et al. Curr Opin Struct Biol. 2004 Feb;14(1):43-9. doi: 10.1016/j.sbi.2004.01.003. Curr Opin Struct Biol. 2004. PMID: 15102448 Review.
Cited by
- Twist-open mechanism of DNA damage recognition by the Rad4/XPC nucleotide excision repair complex.
Velmurugu Y, Chen X, Slogoff Sevilla P, Min JH, Ansari A. Velmurugu Y, et al. Proc Natl Acad Sci U S A. 2016 Apr 19;113(16):E2296-305. doi: 10.1073/pnas.1514666113. Epub 2016 Mar 31. Proc Natl Acad Sci U S A. 2016. PMID: 27035942 Free PMC article. - The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage.
Prakash A, Doublié S, Wallace SS. Prakash A, et al. Prog Mol Biol Transl Sci. 2012;110:71-91. doi: 10.1016/B978-0-12-387665-2.00004-3. Prog Mol Biol Transl Sci. 2012. PMID: 22749143 Free PMC article. Review. - Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA.
Imamura K, Averill A, Wallace SS, Doublié S. Imamura K, et al. J Biol Chem. 2012 Feb 3;287(6):4288-98. doi: 10.1074/jbc.M111.315309. Epub 2011 Dec 14. J Biol Chem. 2012. PMID: 22170059 Free PMC article. - Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Lee SJ, Sung RJ, Verdine GL. Lee SJ, et al. Research (Wash D C). 2019 Aug 28;2019:5641746. doi: 10.34133/2019/5641746. eCollection 2019. Research (Wash D C). 2019. PMID: 31549070 Free PMC article. - Detection of damaged DNA bases by DNA glycosylase enzymes.
Friedman JI, Stivers JT. Friedman JI, et al. Biochemistry. 2010 Jun 22;49(24):4957-67. doi: 10.1021/bi100593a. Biochemistry. 2010. PMID: 20469926 Free PMC article. Review.
References
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
- Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. - PubMed
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. - PubMed
- Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. - PubMed
- Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA100742-06A1/CA/NCI NIH HHS/United States
- P01 GM047467-100006/GM/NIGMS NIH HHS/United States
- CA100742/CA/NCI NIH HHS/United States
- R01 GM044853/GM/NIGMS NIH HHS/United States
- R01 CA100742/CA/NCI NIH HHS/United States
- R01 GM044853-18/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM030804/GM/NIGMS NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- R01 GM030804/GM/NIGMS NIH HHS/United States
- GM044853/GM/NIGMS NIH HHS/United States
- GM047467/GM/NIGMS NIH HHS/United States
- P01 GM047467/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials