Nix is a selective autophagy receptor for mitochondrial clearance - PubMed (original) (raw)
doi: 10.1038/embor.2009.256. Epub 2009 Dec 11.
Vladimir Kirkin, David G McEwan, Ji Zhang, Philipp Wild, Alexis Rozenknop, Vladimir Rogov, Frank Löhr, Doris Popovic, Angelo Occhipinti, Andreas S Reichert, Janos Terzic, Volker Dötsch, Paul A Ney, Ivan Dikic
Affiliations
- PMID: 20010802
- PMCID: PMC2816619
- DOI: 10.1038/embor.2009.256
Nix is a selective autophagy receptor for mitochondrial clearance
Ivana Novak et al. EMBO Rep. 2010 Jan.
Abstract
Autophagy is the cellular homeostatic pathway that delivers large cytosolic materials for degradation in the lysosome. Recent evidence indicates that autophagy mediates selective removal of protein aggregates, organelles and microbes in cells. Yet, the specificity in targeting a particular substrate to the autophagy pathway remains poorly understood. Here, we show that the mitochondrial protein Nix is a selective autophagy receptor by binding to LC3/GABARAP proteins, ubiquitin-like modifiers that are required for the growth of autophagosomal membranes. In cultured cells, Nix recruits GABARAP-L1 to damaged mitochondria through its amino-terminal LC3-interacting region. Furthermore, ablation of the Nix:LC3/GABARAP interaction retards mitochondrial clearance in maturing murine reticulocytes. Thus, Nix functions as an autophagy receptor, which mediates mitochondrial clearance after mitochondrial damage and during erythrocyte differentiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
Nix interacts with Atg8/LC3/GABARAP proteins. (A) Yeast clones harbouring empty bait vector (pYTH9, mock) or those encoding ScAtg8, LC3B, Atg12 and SUMO1 were transformed with empty prey vector (pACT2, mock) or those encoding fragments of ATG4A, ATG4B, NBR1, p62 and full-length Nix. Interaction was assessed by yeast growth on the SD-W/-L/-H medium with a β-gal assay. (B) GST pulldown assays using cell extracts of Flag-Nix-expressing COS7 cells and immobilized GST or the indicated GST fusions. Coprecipitated Nix was detected with Flag antibodies. (C) Purified GST-tagged NixΔTM was cleaved from the GST moiety by thrombin, purified and used for precipitation by GST fusion proteins. Precipitated proteins were analysed by western blotting (WB) with Nix antibodies. Ponceau S staining was used to visualize GST-fusion proteins. (D) HeLa cells were treated as indicated and endogenous proteins were immunoprecipitated (IP) from cell extracts using Nix antibody and analysed by using Nix and LC3 antibodies. β-gal, β-galactosidase; BafA1, bafilomycin-A1; CCCP, carbonyl cyanide m-chlorophenyl hidrazone; GST, glutathione-_S_-transferase; IgG, immunoglobulin G; NixΔTM, recombinant Nix lacking a TM; SUMO1, small ubiquitin-like modifier 1; TCL, total cell lysate; TM, transmembrane domain.
Figure 2
Identification of Nix-LIRs. (A) Amino-acid sequences of Nix and related BNIP3 protein from the indicated species were aligned using the ClustalW2 program (EMBL-EBI, Cambridge, UK). The published LIR sequences were aligned manually alongside Nix/BNIP3 for comparison. The shaded regions indicate highly conserved residues in the two putative LIRs. Consensus symbols * (identical residues), : (conserved substitution) and . (semi-conserved substitution) are valid for the Nix/BNIP3 alignment only. (B) A schematic of Nix showing its domain organization and the point mutations used in this study to identify LIRs. (C) Lysates of COS7 cells transfected with Flag-wt-Nix or the indicated mutants were incubated with immobilized GST-LC3A. The coprecipitated proteins were detected by western blotting (WB) with Flag antibodies. BNIP3, Bcl2/E1B 19kDa-interacting protein 3-like protein; Dr, Danio reio; GST, glutathione-_S_-transferase; Hs, Homo sapiens; LIR, LC3-interacting region; Mm, Mus musculus; wt, wild type; Xl, Xenopus laevis.
Figure 3
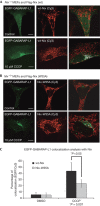
Nix recruits EGFP-GABARAP-L1 to stressed mitochondria in reconstituted _Nix_−/− MEFs. _Nix_−/− MEFs cotransfected with EGFP-GABARAP-L1 and wt-Nix (A), or Nix-W35A (B), were incubated with and without mitochondrial poison CCCP for 3 h. Cells were then fixed and analysed for colocalization between EGFP-GABARAP-L1 and Nix (stained with Flag antibodies). The final panel in each row shows colocalization between Flag-Nix-Cy3 and GABARAP-L1 using the ‘colocalization' highlighter plug-in for ImageJ software (NIH, Bethesda, MA, USA). (C) Quantification was achieved by counting the number of complete colocalizations, EGFP/Cy3 per cell for approximately 30 cells per condition. Results are expressed as a percentage of colocalization (EGFP:Cy3) in control and CCCP-treated cells. Error bars in (C) show the standard deviation (s.d.) of three independent experiments. Statistical analysis was performed using the Mann–Whitney _t_-test. CCCP, carbonyl cyanide m-chlorophenyl hidrazone; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescent protein; MEF, mouse embryonic fibroblast; wt, wild type.
Figure 4
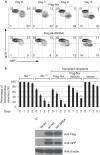
Mitochondrial clearance is impaired in _Nix_−/− reticulocytes transduced with Nix-W35A. (A) Flow cytometry of reticulocyte-enriched blood, cultured in vitro for 3 days, stained with MTR. The blood was taken from mice transplanted with _Nix_−/− bone marrow and reconstituted with wt-Nix and Nix-W35A viruses. Reticulocytes were induced by phenylhydrazine treatment. The viruses also express GFP. (B) Mitochondrial clearance in the GFP-positive fraction of the reticulocyte-enriched blood, as described in panel (A). Mice were reconstituted with wt-Nix, LIR-W35A or empty vector viruses. Data from _Nix_−/− and Nix+/+ mice are shown for comparison. (C) Anti-Flag, anti-GFP and anti-β-actin western blots of fetal liver cells transduced with Flag-Nix and Flag-W35A-Nix viruses, normalized to total protein. GFP, green fluorescent protein; LIR, LC3-interacting region; MTR, MitoTracker Red; wt, wild type.
Comment in
- Opening a new DOR to autophagy.
Spowart J, Lum JJ. Spowart J, et al. EMBO Rep. 2010 Jan;11(1):4-5. doi: 10.1038/embor.2009.265. EMBO Rep. 2010. PMID: 20033084 Free PMC article. No abstract available.
Similar articles
- Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Rogov VV, Suzuki H, Marinković M, Lang V, Kato R, Kawasaki M, Buljubašić M, Šprung M, Rogova N, Wakatsuki S, Hamacher-Brady A, Dötsch V, Dikic I, Brady NR, Novak I. Rogov VV, et al. Sci Rep. 2017 Apr 25;7(1):1131. doi: 10.1038/s41598-017-01258-6. Sci Rep. 2017. PMID: 28442745 Free PMC article. - FKBP8 recruits LC3A to mediate Parkin-independent mitophagy.
Bhujabal Z, Birgisdottir ÅB, Sjøttem E, Brenne HB, Øvervatn A, Habisov S, Kirkin V, Lamark T, Johansen T. Bhujabal Z, et al. EMBO Rep. 2017 Jun;18(6):947-961. doi: 10.15252/embr.201643147. Epub 2017 Apr 5. EMBO Rep. 2017. PMID: 28381481 Free PMC article. - Dimerization of mitophagy receptor BNIP3L/NIX is essential for recruitment of autophagic machinery.
Marinković M, Šprung M, Novak I. Marinković M, et al. Autophagy. 2021 May;17(5):1232-1243. doi: 10.1080/15548627.2020.1755120. Epub 2020 Apr 24. Autophagy. 2021. PMID: 32286918 Free PMC article. - A brief overview of BNIP3L/NIX receptor-mediated mitophagy.
Marinković M, Novak I. Marinković M, et al. FEBS Open Bio. 2021 Dec;11(12):3230-3236. doi: 10.1002/2211-5463.13307. Epub 2021 Oct 11. FEBS Open Bio. 2021. PMID: 34597467 Free PMC article. Review. - Mechanisms and biology of B-cell leukemia/lymphoma 2/adenovirus E1B interacting protein 3 and Nip-like protein X.
Zhang J, Ney PA. Zhang J, et al. Antioxid Redox Signal. 2011 May 15;14(10):1959-69. doi: 10.1089/ars.2010.3772. Epub 2011 Mar 4. Antioxid Redox Signal. 2011. PMID: 21126215 Free PMC article. Review.
Cited by
- Mitochondrial Quality Control Orchestrates the Symphony of B Cells and Plays Critical Roles in B Cell-Related Diseases.
Li W, Cai P, Xu Y, Tian W, Jing L, Lv Q, Zhao Y, Wang H, Shao Q. Li W, et al. J Immunol Res. 2024 Oct 17;2024:5577506. doi: 10.1155/2024/5577506. eCollection 2024. J Immunol Res. 2024. PMID: 39449998 Free PMC article. Review. - Autophagy adaptors mediate Parkin-dependent mitophagy by forming sheet-like liquid condensates.
Yang Z, Yoshii SR, Sakai Y, Zhang J, Chino H, Knorr RL, Mizushima N. Yang Z, et al. EMBO J. 2024 Nov;43(22):5613-5634. doi: 10.1038/s44318-024-00272-5. Epub 2024 Oct 17. EMBO J. 2024. PMID: 39420095 Free PMC article. - Mitophagy-associated programmed neuronal death and neuroinflammation.
Zhu Y, Zhang J, Deng Q, Chen X. Zhu Y, et al. Front Immunol. 2024 Oct 2;15:1460286. doi: 10.3389/fimmu.2024.1460286. eCollection 2024. Front Immunol. 2024. PMID: 39416788 Free PMC article. Review. - GABA(A) Receptor Activation Drives GABARAP-Nix Mediated Autophagy to Radiation-Sensitize Primary and Brain-Metastatic Lung Adenocarcinoma Tumors.
Bhattacharya D, Barrile R, Toukam DK, Gawali VS, Kallay L, Ahmed T, Brown H, Rezvanian S, Karve A, Desai PB, Medvedovic M, Wang K, Ionascu D, Harun N, Vallabhapurapu S, Wang C, Qi X, Baschnagel AM, Kritzer JA, Cook JM, Pomeranz Krummel DA, Sengupta S. Bhattacharya D, et al. Cancers (Basel). 2024 Sep 15;16(18):3167. doi: 10.3390/cancers16183167. Cancers (Basel). 2024. PMID: 39335139 Free PMC article. - Melatonin regulates mitochondrial dynamics and mitophagy: Cardiovascular protection.
Rahmani S, Roohbakhsh A, Pourbarkhordar V, Hayes AW, Karimi G. Rahmani S, et al. J Cell Mol Med. 2024 Sep;28(18):e70074. doi: 10.1111/jcmm.70074. J Cell Mol Med. 2024. PMID: 39333694 Free PMC article. Review.
References
- Bocharov EV et al. (2007) Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J Biol Chem 282: 16256–16266 - PubMed
- Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB (2007) Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci 120 (Part 23): 4155–4166 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases